For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MASLRLTTKVQVSGRRFLVRRLEHAIVRRDTQMFDDPLQFYSRSASLGIVVAVLILAGAVMLAYVKPQGQ LGDTNLVADRASDQLYVTVSGQLHPVYNLTSARLVLGNPAAPTSMRSSELNKLPKGQTIGIPGAPYATPV SADSASTWSLCDTVAYAGTANPVTRTALIAMPLQIDAAIAPLRANEALLVSYRHSTWIVTAQGRHAIDLS DRALTTAMGIGETATPTPISEGMFNALPDSGPWRLPSIDAAGAPNTLGLPAALVIGSVFRVETTAGTKHF VVLRDGVAPVNDDTAGALRDMESYGLIDPPPVPPDLVVDLPQRVYQSPLPDEPLKIISRVQDPTLCWSWQ RSAGDQAPTTSVLSGRHLPLPPSVVSAGISQIRGTATIYIDGGKFLVLQSPDPRYAESMYYVDPQGVRYG VPDAAVANALGLGSPQDAPWQVVRLLVDGPVLSKDAALLEHETLPPDPNPAKIDAAAAGAP
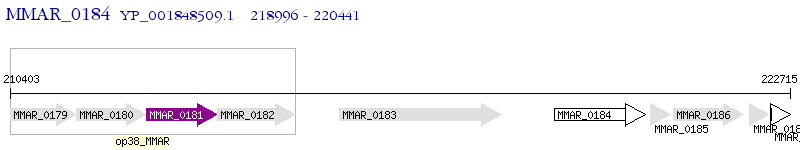
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_0184 | - | - | 100% (481) | hypothetical protein MMAR_0184 |
| M. marinum M | MMAR_5444 | - | 0.0 | 70.29% (478) | hypothetical protein MMAR_5444 |
| M. marinum M | MMAR_0542 | - | 4e-75 | 35.14% (498) | hypothetical protein MMAR_0542 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3899 | - | 0.0 | 72.59% (478) | hypothetical protein Mb3899 |
| M. gilvum PYR-GCK | Mflv_0772 | - | 1e-170 | 59.83% (478) | hypothetical protein Mflv_0772 |
| M. tuberculosis H37Rv | Rv3869 | - | 0.0 | 72.59% (478) | hypothetical protein Rv3869 |
| M. leprae Br4923 | MLBr_00054 | - | 0.0 | 65.13% (476) | hypothetical protein MLBr_00054 |
| M. abscessus ATCC 19977 | MAB_3759c | - | 2e-69 | 34.77% (486) | hypothetical protein MAB_3759c |
| M. avium 104 | MAV_4870 | - | 3e-77 | 34.71% (510) | hypothetical protein MAV_4870 |
| M. smegmatis MC2 155 | MSMEG_0060 | - | 1e-170 | 60.42% (475) | hypothetical protein MSMEG_0060 |
| M. thermoresistible (build 8) | TH_3503 | - | 1e-170 | 60.04% (483) | PUTATIVE protein of unknown function DUF690 |
| M. ulcerans Agy99 | MUL_1205 | - | 3e-74 | 34.74% (498) | hypothetical protein MUL_1205 |
| M. vanbaalenii PYR-1 | Mvan_0073 | - | 1e-169 | 58.68% (484) | hypothetical protein Mvan_0073 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_0772|M.gilvum_PYR-GCK -------------------------------MAGFRLTTKVQVSGWRFLL
Mvan_0073|M.vanbaalenii_PYR-1 -------------------------------MAGFRLTTKVQVSGWRFLL
TH_3503|M.thermoresistible__bu -------------------------------MAGFRLTTKVQVSGWRFLL
MSMEG_0060|M.smegmatis_MC2_155 -------------------------------MAGFRLTTKVQVSGWRFLL
Mb3899|M.bovis_AF2122/97 --------------------------------MGLRLTTKVQVSGWRFLL
Rv3869|M.tuberculosis_H37Rv --------------------------------MGLRLTTKVQVSGWRFLL
MLBr_00054|M.leprae_Br4923 --------------------------------MGLRLTTKVQVSGWRFLL
MMAR_0184|M.marinum_M -------------------------------MASLRLTTKVQVSGRRFLV
MAV_4870|M.avium_104 MTANNDPDD-----RRSFTSRTPVNDNPDQVQYRRGFVTRHQVSGWRFVM
MUL_1205|M.ulcerans_Agy99 MTTN-EPDGEWAGARRSFTSRTPVNDNPDKVEYRRGFVTRHQVTGWRFVM
MAB_3759c|M.abscessus_ATCC_199 ------------------MARQPT--------------TRLQVSGYRFLV
*: **:* **::
Mflv_0772|M.gilvum_PYR-GCK RRVEHAIVRRDTRMFDDPLQFYSRAVSAGIVIAVLICLGAVLLAYFKPLG
Mvan_0073|M.vanbaalenii_PYR-1 RRVEHAIVRRDTRMFDDPLQFYSRAVTTGIVIAVLICLGAVLLAYFKPLG
TH_3503|M.thermoresistible__bu RRVEHAIVRRDTRMFDDPLQFYSRAVQAGIVISVLICLGAVLLAYFKPLG
MSMEG_0060|M.smegmatis_MC2_155 RRVEHAIVRRDTRMFDDPLQFYSRAVFAGVVVSVLICLGAALMAYFKPLG
Mb3899|M.bovis_AF2122/97 RRLEHAIVRRDTRMFDDPLQFYSRSIALGIVVAVLILAGAALLAYFKPQG
Rv3869|M.tuberculosis_H37Rv RRLEHAIVRRDTRMFDDPLQFYSRSIALGIVVAVLILAGAALLAYFKPQG
MLBr_00054|M.leprae_Br4923 RRVEHAIVRRDTRMFDDPLQFYSRSIALGIVVAVALLIGAVLLAYFKPQG
MMAR_0184|M.marinum_M RRLEHAIVRRDTQMFDDPLQFYSRSASLGIVVAVLILAGAVMLAYVKPQG
MAV_4870|M.avium_104 RRIASGIALHDTRMLVDPLRTQSRAVAMGAVLLVTGLAGCFVFSLIRPNG
MUL_1205|M.ulcerans_Agy99 RRIASGIALHDTRMLVDPLRTQSRSVLMGVLIFITGLVGCFVFSLIRPNG
MAB_3759c|M.abscessus_ATCC_199 RRMEHALVRRDVRMIHDPMRSQSSSLMVGIVLATVVVAGCAILAFFRPNM
**: .:. :*.:*: **:: * : * :: *. ::: .:*
Mflv_0772|M.gilvum_PYR-GCK KRGDDTLLVDRTTNQLYLVLPDSSQLRPVYNLTSARLILGEGKTPVAVKS
Mvan_0073|M.vanbaalenii_PYR-1 KRGGDTLLVDRTTNQLFIVLPDSGQLRPVYNLTSARLILGDGKTPVAVKS
TH_3503|M.thermoresistible__bu KRGGDTLLVDRTTNQLYVTLPGSDQLRPVYNLTSARLVLANNATPSAVKS
MSMEG_0060|M.smegmatis_MC2_155 KQGSDQLLVDRTTNQLYVMLPGSNQLRPVYNLTSARLVLGNASNPVAVKS
Mb3899|M.bovis_AF2122/97 KLGGTSLFTDRATNQLYVLLSG--QLHPVYNLTSARLVLGNPANPATVKS
Rv3869|M.tuberculosis_H37Rv KLGGTSLFTDRATNQLYVLLSG--QLHPVYNLTSARLVLGNPANPATVKS
MLBr_00054|M.leprae_Br4923 KLGASTLLTDRATNQLYVLLSG--HLYPVYNLTSARLALGKPANPAAVKS
MMAR_0184|M.marinum_M QLGDTNLVADRASDQLYVTVSG--QLHPVYNLTSARLVLGNPAAPTSMRS
MAV_4870|M.avium_104 TAGTNAVLADRSTAALYVRVGD--DLHPVLNLTSARLIVGKPVNPTTVKS
MUL_1205|M.ulcerans_Agy99 QVGSNAVLADRSTAALYVRVGD--TLHPVLNLTSARLIVGHPVNPTTVKS
MAB_3759c|M.abscessus_ATCC_199 PLGDQPIVMGKDTGAVYVRIQD--TLHPALNLASARLISGNNENPKPVKD
* :. .: : ::: : . * *. **:**** .. * .::.
Mflv_0772|M.gilvum_PYR-GCK DELDRTPKGLPVGIPGAPYATA-VSAPSESQWSLCDTVVKPD----RVAP
Mvan_0073|M.vanbaalenii_PYR-1 QELDKMPKGQPVGIPGAPYATP-VASSAESQWTLCDTVIKPD----SVAP
TH_3503|M.thermoresistible__bu EELDRMPKGQPIGIPGAPYATP-LAAP-QSTWTLCDTVTRPE----SVSP
MSMEG_0060|M.smegmatis_MC2_155 EELNRISKGQSIGIPGAPYATP-TGTP-ASQWTLCDTVAKPD----SSAP
Mb3899|M.bovis_AF2122/97 SELSKLPMGQTVGIPGAPYATP-VSAGSTSIWTLCDTVARAD----STSP
Rv3869|M.tuberculosis_H37Rv SELSKLPMGQTVGIPGAPYATP-VSAGSTSIWTLCDTVARAD----STSP
MLBr_00054|M.leprae_Br4923 SELTKLPIGQTIGIPGAPYATP-VSGDSSSTWTLCDTVAKIE----SESP
MMAR_0184|M.marinum_M SELNKLPKGQTIGIPGAPYATP-VSADSASTWSLCDTVAYAG----TANP
MAV_4870|M.avium_104 AELDRFPRGNLLGIPGAPERMVQNPS-TDANWTVCDSVSEPAPGGHGAHS
MUL_1205|M.ulcerans_Agy99 TELDRFPRGNLIGIPGAPERMVQSEA-RDSDWTVCDAISSGSQGGSAAHS
MAB_3759c|M.abscessus_ATCC_199 SELAKARRGPLVGIPGAPASIPGALTGDDTIWTVCDVATPPP-----SST
** : * :****** : *::** .
Mflv_0772|M.gilvum_PYR-GCK AVESSVLVTPLALDASVGAMRPEQGMLVTYKDRTWLVTPQGRHLIDMADR
Mvan_0073|M.vanbaalenii_PYR-1 AVESSVLITPLALDASVGAMRPEQGMLVSFRDQNWLVTEAGRHLIDLADR
TH_3503|M.thermoresistible__bu TLRTSVLVLPLATDTAVGPMNPNQGMLVTYQGKPWLVTDRGRHSIDLADR
MSMEG_0060|M.smegmatis_MC2_155 KVETSILIRTLAIDSGVGPIRADQGMLVSYEGANWLITEGGRHSIDLADR
Mb3899|M.bovis_AF2122/97 VVQTAVIAMPLEIDASIDPLQSHEAVLVSYQGETWIVTTKGRHAIDLTDR
Rv3869|M.tuberculosis_H37Rv VVQTAVIAMPLEIDASIDPLQSHEAVLVSYQGETWIVTTKGRHAIDLTDR
MLBr_00054|M.leprae_Br4923 AVQTSVIARSLQIDPAINPLQPNEALLASYRDKTWLVNSKGRHSIDLADR
MMAR_0184|M.marinum_M VTRTALIAMPLQIDAAIAPLRANEALLVSYRHSTWIVTAQGRHAIDLSDR
MAV_4870|M.avium_104 TGVTVIAGRPDSSGARAAALPSQQAILAERDGGTWLLWNGRRSQINLADH
MUL_1205|M.ulcerans_Agy99 TGVTVIAGSPEPSGHRATALGAGQVILVNDGTSSWLLWDGKRSRMDLADH
MAB_3759c|M.abscessus_ATCC_199 VTTTVIAGKVTLENG-NQEMPKDRYALVTVNSQMYLLYDGKRAAVDLTNP
: : . : . *. ::: * :::::
Mflv_0772|M.gilvum_PYR-GCK AVTSAVGIPVT----ARAAPISEGLFNALPDEGPWRLPDIPGAGTPNTVG
Mvan_0073|M.vanbaalenii_PYR-1 AVTSAVGIPVT----ARSAPISEGLFNALPNAGPWRLPDIPAGGSPNSVG
TH_3503|M.thermoresistible__bu AVTSAVGIPVT----ARATPISTGLFNALPNAGPWQVPDVPAAGAPNTVG
MSMEG_0060|M.smegmatis_MC2_155 AVTSAVGIPVT----AKPTPISQGLFNALPNRGPWQLPQIPAAGAPNSVG
Mb3899|M.bovis_AF2122/97 ALTSSMGIPVT----ARPTPISEGMFNALPDMGPWQLPPIPAAGAPNSLG
Rv3869|M.tuberculosis_H37Rv ALTSSMGIPVT----ARPTPISEGMFNALPDMGPWQLPPIPAAGAPNSLG
MLBr_00054|M.leprae_Br4923 ALTSAIGISVN----VKPTPLSEGLFNALPDVGRWELPTIPDTGAPNSLG
MMAR_0184|M.marinum_M ALTTAMGIGET----ATPTPISEGMFNALPDSGPWRLPSIDAAGAPNTLG
MAV_4870|M.avium_104 AVTNALGLTENNSELPAPRPIALGLFNAIPEAPSLSAPGIPNAGAPAPFA
MUL_1205|M.ulcerans_Agy99 AVTNALGFA---TDLPAPRPIAPGLFNAIPEAAPLAAPAIPDAGNPAAFA
MAB_3759c|M.abscessus_ATCC_199 AVTRALRLEG-----VTPRPISMNVLNAIPEVPAIAPPPIPDAGAASPIT
*:* :: : . *:: .::**:*: * : * . ..
Mflv_0772|M.gilvum_PYR-GCK LPPALVIGSVFVTVTDMGEQTDEQGYVVLPSGVAKVNSTTAAALRATNSY
Mvan_0073|M.vanbaalenii_PYR-1 LPPELVVGSVFVTVTDAGEQAGEQNYVVLPNGVAKVNTTTAAALRATNSF
TH_3503|M.thermoresistible__bu LPEHLVIGSVFKTVTDS-----EQHYVVLPDGVARVNNTTAAALRARNSY
MSMEG_0060|M.smegmatis_MC2_155 LPENLVIGSVFRTATES----DPQHYVVLPDGVARVNNTTAAALRATNSY
Mb3899|M.bovis_AF2122/97 LPDDLVIGSVFQIHTDK----GPQYYVVLPDGIAQVNATTAAALRATQAH
Rv3869|M.tuberculosis_H37Rv LPDDLVIGSVFQIHTDK----GPQYYVVLPDGIAQVNATTAAALRATQAH
MLBr_00054|M.leprae_Br4923 LSPDLVIGSVFQIQMEK----SPQYYVVLSDGIAQVNATTADALRATQSH
MMAR_0184|M.marinum_M LPAALVIGSVFRVETTA----GTKHFVVLRDGVAPVNDDTAGALRDMESY
MAV_4870|M.avium_104 VPAPIGAVVVSYTLDQSSS-GAPRYYAVLPDGLQQISPVLAAILRNTNSY
MUL_1205|M.ulcerans_Agy99 VPGPIGSVVQAYTLGDNS----IQYYAVLPDGLQPISPVLAAIVRNTNSY
MAB_3759c|M.abscessus_ATCC_199 TAGVRVGSVIKMSRADSA-----EYYVVLAGGVQRINEVTADLIRFSDSQ
. . :.** .*: :. * :* ::
Mflv_0772|M.gilvum_PYR-GCK GLIEPPSMESSVVARIPEQ--VYTSPLPDRPMDVLGRQEVPTLCWSWQ--
Mvan_0073|M.vanbaalenii_PYR-1 GLIEPPSLESSAVARIPEQ--VYDSPLPGVPMDVLARQELPTLCWAWQ--
TH_3503|M.thermoresistible__bu GLVMPPTIESSVVARIPEH--VFVSPLPDRPLDMLLREETPTLCWSWQ--
MSMEG_0060|M.smegmatis_MC2_155 GLMQPPAVEASVVAKIPEQ--VYVSPLPDQPLDVLLRQDSPVLCWSWQ--
Mb3899|M.bovis_AF2122/97 GLVAPPAMVPSLVVRIAER--VYPSPLPDEPLKIVSRPQDPALCWSWQ--
Rv3869|M.tuberculosis_H37Rv GLVAPPAMVPSLVVRIAER--VYPSPLPDEPLKIVSRPQDPALCWSWQ--
MLBr_00054|M.leprae_Br4923 GLVAPPSLVPNVVVQIPER--VYDSPLPDEPLKMVARDDYPTLCWAWE--
MMAR_0184|M.marinum_M GLIDPPPVPPDLVVDLPQR--VYQSPLPDEPLKIISRVQDPTLCWSWQ--
MAV_4870|M.avium_104 GLDQPPRLGADAVAKLPVSRMLDTARYPDQHVSLVDTAKSPVTCAYWS--
MUL_1205|M.ulcerans_Agy99 GLQQPPRIGADDVAKLPVSRMLDTTRYPATPMHLVDTESNPITCAYWS--
MAB_3759c|M.abscessus_ATCC_199 GAQEIVTVAPDVIAGHPSLNLLAVQTFPDRAGKIVDVPEKSVLCSSWKPG
* : .. :. . : * :: . . * *.
Mflv_0772|M.gilvum_PYR-GCK RVAGDQAPKMTVIAGRRLPLPPSALATGIDQISG------DATVYVDGGQ
Mvan_0073|M.vanbaalenii_PYR-1 RVAGDQAPKMTVIAGRRLPLPPSALSTGIDQITG------DATVYIDGGQ
TH_3503|M.thermoresistible__bu REPGDQAPETTVIAGRRLPIAPSAMNRGIDQIGG------DATVYLAGGQ
MSMEG_0060|M.smegmatis_MC2_155 REPGDQAPKTTVIAGRRLPLPANAIGTGIDQIGG------DSTVYIEGGQ
Mb3899|M.bovis_AF2122/97 RSAGDQSPQSTVLSGRHLPISPSAMNMGIKQIHG------TATVYLDGGK
Rv3869|M.tuberculosis_H37Rv RSAGDQSPQSTVLSGRHLPISPSAMNMGIKQIHG------TATVYLDGGK
MLBr_00054|M.leprae_Br4923 RKASDQAPKRTMLIGQHLPLQPSAMSTGIKQIRG------TATVYIDGGR
MMAR_0184|M.marinum_M RSAGDQAPTTSVLSGRHLPLPPSVVSAGISQIRG------TATIYIDGGK
MAV_4870|M.avium_104 KPAGAATSSLNLLSGSALPIAESIRPVDLVGGGSPQNGPVATRVALPPGT
MUL_1205|M.ulcerans_Agy99 KPVGATTSSLTLLSGAALPVPGAVHTVDLVGGGS-QNGAVATRVALAPGT
MAB_3759c|M.abscessus_ATCC_199 QEKTQEHASTKLLMGSSLPLKTDQVPVVLAQADK--EGPNLDTAYIPPAR
: . .:: * **: : : .
Mflv_0772|M.gilvum_PYR-GCK YIQLQSP----EPRYGENLYYIDPQGVRYGLP---------DQETAARLG
Mvan_0073|M.vanbaalenii_PYR-1 YIRLQSP----DPRFGENLYYIDPQGVRYGLP---------DEETAAKLG
TH_3503|M.thermoresistible__bu YIRLQSP----DPRYGESLYYIDPQGVRYGLP---------DEDTAGYLG
MSMEG_0060|M.smegmatis_MC2_155 FVRLQSP----DPRVGESMYYIDPQGVRYGIA---------NDDAAKNLG
Mb3899|M.bovis_AF2122/97 FVALQSP----DPRYTESMYYIDPQGVRYGVP---------NAETAKSLG
Rv3869|M.tuberculosis_H37Rv FVALQSP----DPRYTESMYYIDPQGVRYGVP---------NAETAKSLG
MLBr_00054|M.leprae_Br4923 YVALQSP----DPRYSESLYYVDPEGVRYGLA---------NSEAVKALG
MMAR_0184|M.marinum_M FLVLQSP----DPRYAESMYYVDPQGVRYGVP---------DAAVANALG
MAV_4870|M.avium_104 GYFTQTASGGAGSPGAGSLFWVSDTGVRYGIDNEAEHSAAGHGKTVEALG
MUL_1205|M.ulcerans_Agy99 GYFTQTAGGGPAAPPTGSLFWVSDTGVRYGIDTEGE----GGRKTVEALG
MAB_3759c|M.abscessus_ATCC_199 SAFVRSTGLGGDTGRAESLYLILDTGVRFGIG---------NLEAAKALG
::. . .:: : ***:*: .. **
Mflv_0772|M.gilvum_PYR-GCK LS-APRTAPWQVVSLLVDGPVLSQQAALIEHDTLPPNPNPRRAGGGSTA-
Mvan_0073|M.vanbaalenii_PYR-1 LS-SPRTAPWQVVSLLVDGPVLSQQAALVEHDTLPPNPNPRSVGADAAAP
TH_3503|M.thermoresistible__bu LH-GPATAPWQVVSLLVDGPVLSKAAALVEHDVLPGNPHPRKIADGAAA-
MSMEG_0060|M.smegmatis_MC2_155 LA-GPVNAPWQVVGLLVDGPVLSKEAALIEHDTLPADPNPRKVASGEG--
Mb3899|M.bovis_AF2122/97 LS-SPQNAPWEIVRLLVDGPVLSKDAALLEHDTLPADPSPRKVPAGASG-
Rv3869|M.tuberculosis_H37Rv LS-SPQNAPWEIVRLLVDGPVLSKDAALLEHDTLPADPSPRKVPAGASG-
MLBr_00054|M.leprae_Br4923 LA-SPQTAPWVIVRLLVEGPVLSRDAALLEHDTLIADPSPRKVPAGYSG-
MMAR_0184|M.marinum_M LG-SPQDAPWQVVRLLVDGPVLSKDAALLEHETLPPDPNPAKIDAAAAG-
MAV_4870|M.avium_104 LSGSPLPIPWSVLSLFAKGPTLSRADALLAHDGLPPDQAPGRVAAAAPQA
MUL_1205|M.ulcerans_Agy99 LQDPPVLIPWSVLSLFAPGPTLSRADALMAHDGLAPDSKPGRPAS----A
MAB_3759c|M.abscessus_ATCC_199 LNVEPVPAPWPIVGLLPPGPRLSKEDALVVHDGMPPD-KTGLAINPSTSA
* * ** :: *: ** **: **: *: : : .
Mflv_0772|M.gilvum_PYR-GCK -QANVGGGG
Mvan_0073|M.vanbaalenii_PYR-1 VGANNGGGG
TH_3503|M.thermoresistible__bu --APGPGGG
MSMEG_0060|M.smegmatis_MC2_155 ---------
Mb3899|M.bovis_AF2122/97 --AP-----
Rv3869|M.tuberculosis_H37Rv --AP-----
MLBr_00054|M.leprae_Br4923 --VRP----
MMAR_0184|M.marinum_M --AP-----
MAV_4870|M.avium_104 EGAPR----
MUL_1205|M.ulcerans_Agy99 EGGPR----
MAB_3759c|M.abscessus_ATCC_199 TQAPS----