For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
ARQPTTRLQVSGYRFLVRRMEHALVRRDVRMIHDPMRSQSSSLMVGIVLATVVVAGCAILAFFRPNMPLG DQPIVMGKDTGAVYVRIQDTLHPALNLASARLISGNNENPKPVKDSELAKARRGPLVGIPGAPASIPGAL TGDDTIWTVCDVATPPPSSTVTTTVIAGKVTLENGNQEMPKDRYALVTVNSQMYLLYDGKRAAVDLTNPA VTRALRLEGVTPRPISMNVLNAIPEVPAIAPPPIPDAGAASPITTAGVRVGSVIKMSRADSAEYYVVLAG GVQRINEVTADLIRFSDSQGAQEIVTVAPDVIAGHPSLNLLAVQTFPDRAGKIVDVPEKSVLCSSWKPGQ EKTQEHASTKLLMGSSLPLKTDQVPVVLAQADKEGPNLDTAYIPPARSAFVRSTGLGGDTGRAESLYLIL DTGVRFGIGNLEAAKALGLNVEPVPAPWPIVGLLPPGPRLSKEDALVVHDGMPPDKTGLAINPSTSATQA PS*
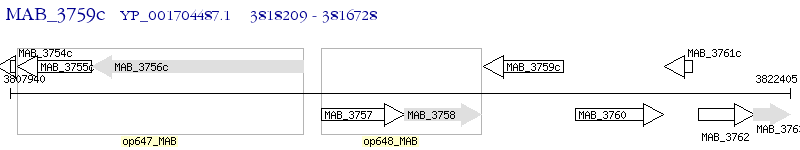
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_3759c | - | - | 100% (493) | hypothetical protein MAB_3759c |
| M. abscessus ATCC 19977 | MAB_2233c | - | 2e-69 | 35.31% (490) | hypothetical protein MAB_2233c |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3480c | - | 1e-111 | 47.27% (476) | hypothetical protein Mb3480c |
| M. gilvum PYR-GCK | Mflv_4962 | - | 1e-105 | 45.99% (474) | hypothetical protein Mflv_4962 |
| M. tuberculosis H37Rv | Rv3450c | - | 1e-111 | 47.27% (476) | hypothetical protein Rv3450c |
| M. leprae Br4923 | MLBr_02536 | - | 7e-76 | 36.04% (505) | hypothetical protein MLBr_02536 |
| M. marinum M | MMAR_1099 | - | 1e-106 | 44.63% (484) | hypothetical protein MMAR_1099 |
| M. avium 104 | MAV_4392 | - | 8e-99 | 43.04% (474) | hypothetical protein MAV_4392 |
| M. smegmatis MC2 155 | MSMEG_1534 | - | 1e-100 | 44.72% (483) | hypothetical protein MSMEG_1534 |
| M. thermoresistible (build 8) | TH_4296 | - | 1e-113 | 48.25% (487) | PUTATIVE putative protein of unknown function DUF690 |
| M. ulcerans Agy99 | MUL_0857 | - | 1e-105 | 44.35% (487) | hypothetical protein MUL_0857 |
| M. vanbaalenii PYR-1 | Mvan_1444 | - | 1e-112 | 47.46% (472) | hypothetical protein Mvan_1444 |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3480c|M.bovis_AF2122/97 -------------------------------MPSPATTWLHVSGYRFLLR
Rv3450c|M.tuberculosis_H37Rv -------------------------------MPSPATTWLHVSGYRFLLR
MMAR_1099|M.marinum_M ----------------------MDAGDSRGWPPGRPTTWLQVSAYRFLVR
MUL_0857|M.ulcerans_Agy99 ----------------------MDAGDSRGWPPGRPTTWLQVSAYRFLVR
MAV_4392|M.avium_104 -------------------------------MPRRPTTWLQVSAYRYLSR
Mflv_4962|M.gilvum_PYR-GCK --------------------------------MGQPVTRLQISGQRFLTR
Mvan_1444|M.vanbaalenii_PYR-1 --------------------------------MRQPTTRLQVSGHRFLMR
TH_4296|M.thermoresistible__bu --------------------------------VGQPTTRLQVSGYRFMLR
MSMEG_1534|M.smegmatis_MC2_155 -------------------------------MARQPVTRLERSGRRFLIR
MAB_3759c|M.abscessus_ATCC_199 -------------------------------MARQPTTRLQVSGYRFLVR
MLBr_02536|M.leprae_Br4923 MTSNELPGEWSGERRSFFSRTPVNDNPDKVVYRRGFVTRHQVTGWRFVMR
.* . :. *:: *
Mb3480c|M.bovis_AF2122/97 RIECALLFGDVCAATGALRARTTSLALGCVLAIVAAMGCAFVALLRPQSA
Rv3450c|M.tuberculosis_H37Rv RIECALLFGDVCAATGALRARTTSLALGCVLAIVAAMGCAFVALLRPQSA
MMAR_1099|M.marinum_M RLECALLRGGLDPADGRPRASAVSLAAGCVLSVVVLVGCVISALLRPQVG
MUL_0857|M.ulcerans_Agy99 RLECALLRGGLDPADGRPRASAVSLAAGCVLSVVVLVGCVISALLRPQVG
MAV_4392|M.avium_104 RLECALLGQDPAAAGMRARGQPAALAAGAVLTAAALAAATLLGLLRPHVA
Mflv_4962|M.gilvum_PYR-GCK RMEHALVRADARMLDDPLRAQSLSLVVGAVLAAIAVAVCAVIAVVRPGGT
Mvan_1444|M.vanbaalenii_PYR-1 RMEHALVRGDARMLDDPLRGQSLSMTAGAVLAVIAVTVCAVLAFIRPGGE
TH_4296|M.thermoresistible__bu RMEHALVRADVRMLDDPLRAQSLALTAGAVLAAVAVAGCAVLALLRPAGR
MSMEG_1534|M.smegmatis_MC2_155 RTMHALVRGDARMIDDPLRAQSVSFAAGCIVTVLAAVVSAVLALVRPGDT
MAB_3759c|M.abscessus_ATCC_199 RMEHALVRRDVRMIHDPMRSQSSSLMVGIVLATVVVAGCAILAFFRPNMP
MLBr_02536|M.leprae_Br4923 RIASGIALHDTRMLVDPLRTQSRSVLVGALLVITGLIGCFVFSFIRPNGA
* .: . * . :. * :: . . ...**
Mb3480c|M.bovis_AF2122/97 LGQAPIVMGRESGALYVRVDDVWHPVLNLASARLIAATNANPQPVSESEL
Rv3450c|M.tuberculosis_H37Rv LGQAPIVMGRESGALYVRVDDVWHPVLNLASARLIAATNANPQPVSESEL
MMAR_1099|M.marinum_M LGDAQIVVGRESGALYVRVGDTWHPVLNLASARLIAGTGANPKPVREAEV
MUL_0857|M.ulcerans_Agy99 LGDAQIVVGRESGALYVRVGDTWHPVLNLASARLIAGTGANPKPVREAEV
MAV_4392|M.avium_104 LDGARVVLARDSGALFVRVGDTWHPVLNLASARLIAASAVDPLTIRQADL
Mflv_4962|M.gilvum_PYR-GCK LGDARIVVVRESGAMFVRLDDVLHPVYNLASARLLLGSPEVPRVVAQRAI
Mvan_1444|M.vanbaalenii_PYR-1 LGDAPLVVVRESGAMYVRIDDTMHPVFNLASARLILGTAAEPRMVTQRAV
TH_4296|M.thermoresistible__bu LDSAQIVMERESGALYVRIDDTWHPTPNLASARLAARTAADPQPVSATAL
MSMEG_1534|M.smegmatis_MC2_155 LSDAPILLARNSGALHVRIGDTVHPVLNLASARLIVQNPADPVLVDDAAI
MAB_3759c|M.abscessus_ATCC_199 LGDQPIVMGKDTGAVYVRIQDTLHPALNLASARLISGNNENPKPVKDSEL
MLBr_02536|M.leprae_Br4923 AGNNAVLADRSTAALYVRVGDELHPVLNLTSARLIVGRSVNPITVKSSEL
. :: :.:.*:.**: * **. **:**** * : :
Mb3480c|M.bovis_AF2122/97 GHTKRGPLLGIPGAPQLLDQPLAGAESAWAICDSD--------NGGSTTV
Rv3450c|M.tuberculosis_H37Rv GHTKRGPLLGIPGAPQLLDQPLAGAESAWAICDSD--------NGGSTTV
MMAR_1099|M.marinum_M IRAKRGPLLGIPGAPQFVGQSLPVQESAWTICDTD--------RGAATTL
MUL_0857|M.ulcerans_Agy99 IRVKRGPLLGIPGAPQFVGQSLPVQESAWTICDTD--------RGAATTL
MAV_4392|M.avium_104 DGSKRGPLLGIPGAPAFLGRPLAAADTAWSLCDSDG-------AGATTTI
Mflv_4962|M.gilvum_PYR-GCK DHAERGARLGIPAAPEQIPTPLSADEAAWSVCDDD---------RGVTTV
Mvan_1444|M.vanbaalenii_PYR-1 DRAEIGPQVGIPGAPGQISPPLQAAESGWTVCDE----------RSGTTV
TH_4296|M.thermoresistible__bu ADAERGPMVGIPGAPGSIGRALPPGQSTWTLCDG----------TTTTLW
MSMEG_1534|M.smegmatis_MC2_155 DAAPRGPLVGIPGAPARIHPPLSADEITWALCEDLA--------SESTTL
MAB_3759c|M.abscessus_ATCC_199 AKARRGPLVGIPGAPASIPGALTGDDTIWTVCDVATPP---PSSTVTTTV
MLBr_02536|M.leprae_Br4923 DRFPRGNLIGIPGAPERMVQNTT-HDANWTVCDVVSGEGGHAAHSMGVTV
* :***.** : : *::*: .
Mb3480c|M.bovis_AF2122/97 VVGPAEDSSA-QVLTAE-QMILVATESGSP--TYLLYGGRRAVVDLADPA
Rv3450c|M.tuberculosis_H37Rv VVGPAEDSSA-QVLTAE-QMILVATESGSP--TYLLYGGRRAVVDLADPA
MMAR_1099|M.marinum_M LVGRPAQQSV-RRLVAE-QTILVATESGSP--AYLLYDGHRAVVDLADPA
MUL_0857|M.ulcerans_Agy99 LVGRPAQQSV-RRLVAE-QTILVATESGSP--AYLLYDGHRAVVDLADPA
MAV_4392|M.avium_104 LLGRPDAASM-RVLGGG-QAMLVAAAAGSP--AYLLHDGHRARVDLADAA
Mflv_4962|M.gilvum_PYR-GCK VAGPPATEGM-TADRH----VLVTPRGQSAAVTYLLHGGRRSRVDLRHPA
Mvan_1444|M.vanbaalenii_PYR-1 TAGPVAGSAM-TPGAS----VLVTPRGGSAALTYLLHGRWRSRVDLRHPA
TH_4296|M.thermoresistible__bu VGEPVAGDRL-GDRRDDPDAVLVRSAGGHG--TYLLHDGARAAVDLRDTA
MSMEG_1534|M.smegmatis_MC2_155 IAGRSGLAPL-RPGRA----VPVAARGAVT--TYLLFDGRKAQVDLREGA
MAB_3759c|M.abscessus_ATCC_199 IAGKVTLENG-NQEMPKDRYALVTVN----SQMYLLYDGKRAAVDLTNPA
MLBr_02536|M.leprae_Br4923 IAGPPDSHGMRAAVLGSAHGVLVDAPSERGGSTWLLWDGKRSEIDLADHA
* :** . :: :** . *
Mb3480c|M.bovis_AF2122/97 VVWALRLQG-----RVPHVVAQSLLNAVPEAPRITAPRIRGGGR-ASVGL
Rv3450c|M.tuberculosis_H37Rv VVWALRLQG-----RVPHVVAQSLLNAVPEAPRITAPRIRGGGR-ASVGL
MMAR_1099|M.marinum_M VRRALRLEG-----RTPYRVSRSLLSAVPEVPPISALRIGHAGEPALAGK
MUL_0857|M.ulcerans_Agy99 VRRALRLEG-----RTPYRVSRSLLSAVPEVPPISVLRIGHAGEPALAGK
MAV_4392|M.avium_104 VVRTLRLEG-----RAPRLVSQSLLEAVPEAAPIAVPRIPGLGG-AAPGL
Mflv_4962|M.gilvum_PYR-GCK VVRALRLDG-----VVPQPVSMELLAALPEAPAIEPPRIPGLGAPAPGAL
Mvan_1444|M.vanbaalenii_PYR-1 VVRALRLDG-----VVPQPVSPALLAAVPEAPAISPPHIPALGSPGPSTL
TH_4296|M.thermoresistible__bu VVRALRLEN-----VEPQSVSDAVLEAIPESPPLTPPGIPAAGTPGPAPL
MSMEG_1534|M.smegmatis_MC2_155 VVRALRLED-----VTPVPVSPTLLDSVPEAPAVRAPVIPDVGAPGPSAV
MAB_3759c|M.abscessus_ATCC_199 VTRALRLEG-----VTPRPISMNVLNAIPEVPAIAPPPIPDAGAASPITT
MLBr_02536|M.leprae_Br4923 VTDALGFGVGFAEVPAPRPIGAGLFNAIPEAPPLKAPVIPNAGATPSFGV
* :* : * :. :: ::** . : * *
Mb3480c|M.bovis_AF2122/97 PGFLVGGVVRITRASG------------DEYYVVLEDGVQRIGQVAADLL
Rv3450c|M.tuberculosis_H37Rv PGFLVGGVVRITRASG------------DEYYVVLEDGVQRIGQVAADLL
MMAR_1099|M.marinum_M LGFSVGTVLSITAGGG------------AAYYVVLAQGVQRIGQVAADLL
MUL_0857|M.ulcerans_Agy99 LGFSVGTVLSITAGGG------------AAYYVVLAQGVQRIGQVAADLL
MAV_4392|M.avium_104 PGFAIGTVLRIIRADG------------DEYYAVLAGGVQRIGRVAADLL
Mflv_4962|M.gilvum_PYR-GCK RNRTVGTVVTVAHAGTGSPRGA------GDHFVVLADGLQRIGEVTADLI
Mvan_1444|M.vanbaalenii_PYR-1 REHPVGTVVRAS--GTDAT---------AEYFVVLAEGLQRIGEVAADLI
TH_4296|M.thermoresistible__bu SRIPVGTVVRLARASAAGGEVGPESGTESDYFVVLADGVQRVGRITADLI
MSMEG_1534|M.smegmatis_MC2_155 NGLPVGSVVRVSRAVPDDSARG------EEFFVVLTGGLQRVDRVVADLI
MAB_3759c|M.abscessus_ATCC_199 AGVRVGSVIKMSRADS------------AEYYVVLAGGVQRINEVTADLI
MLBr_02536|M.leprae_Br4923 RAPIGAVVVSFGLAALGKNPYDS-----VRYYAVLPDGLQPISPVLAAIL
. *: .:.** *:* :. : * ::
Mb3480c|M.bovis_AF2122/97 RFGDSQGSVNVPTVAPDVIRVAPIVNTLPVSAFPDRPPTPVDGSPGRAVT
Rv3450c|M.tuberculosis_H37Rv RFGDSQGSVNVPTVAPDVIRVAPIVNTLPVSAFPDRPPTPVDGSPGRAVT
MMAR_1099|M.marinum_M RFSDSQGAANAIAVAPDVIRAAPIVETLPVTGYPDTMSEPLDAG----AT
MUL_0857|M.ulcerans_Agy99 RFSDSQGAANAIAVAPDVIRAAPIVETLPVTGYPDTVSEPLDAG----VT
MAV_4392|M.avium_104 RLSNPQGGADAVTVAPDALRTAPVVAALPVAGFPDRAPAVSEPA-----A
Mflv_4962|M.gilvum_PYR-GCK RFTDARAGAEIPAVSADIVGALPVVTSLPVATYPDRA----DVT---TAS
Mvan_1444|M.vanbaalenii_PYR-1 RYTDRRPGAEIPTVSADAVGAVPVVTTLPVTTFPDRG----GVT---AAP
TH_4296|M.thermoresistible__bu RFTTAQPHREIATVAAEVIAAVPAVDHLRVATFPDRI----ETR---SAP
MSMEG_1534|M.smegmatis_MC2_155 RFTYPQPDGEPPLLAADVIAGVPHVETVAIPAVPHGL----SQR---TR-
MAB_3759c|M.abscessus_ATCC_199 RFSDSQGAQEIVTVAPDVIAGHPSLNLLAVQTFPDRAGKIVDVP---EKS
MLBr_02536|M.leprae_Br4923 RNINSYGLQQPPRLGADEVDKLPVSRMLDTERYPEQQISLIDAG---YAP
* : :..: : * : *.
Mb3480c|M.bovis_AF2122/97 TLCVTWTP---AQPGAARVAFLAGSGPPVPLGGVPVTLAQADGRG--PAL
Rv3450c|M.tuberculosis_H37Rv TLCVTWTP---AQPGAARVAFLAGSGPPVPLGGVPVTLAQADGRG--PAL
MMAR_1099|M.marinum_M TLCLSWVS---GHASRPDVAFLAGDGLPVPAGARALPLVQADGRG--PAL
MUL_0857|M.ulcerans_Agy99 TLCLSWVS---RHASRPDVAFLAGDGLPVPAGARALPLVQADGRG--PAL
MAV_4392|M.avium_104 TICAGWGD---ARS----VAFLTGAAVPVPPGRAPVTLAQADGRG--PAL
Mflv_4962|M.gilvum_PYR-GCK VVCATWAP--TAGGHGSRTELLTGPKLPPA--PRPIVVAPGR--------
Mvan_1444|M.vanbaalenii_PYR-1 VICAHWNQ--KNGETTSHTSVLVADSAPRR--TRPVTLVQTDGDG--PAV
TH_4296|M.thermoresistible__bu VLCVQWDG--ADG--SPRTSLLLRDSAPDG--AR-VVLAQADGSG--PAV
MSMEG_1534|M.smegmatis_MC2_155 VLCAAWDP--RGN----QTALLFGEAVG----GATLELAQSDSAG--PNI
MAB_3759c|M.abscessus_ATCC_199 VLCSSWKPGQEKTQEHASTKLLMGSSLPLKTDQVPVVLAQADKEG--PNL
MLBr_02536|M.leprae_Br4923 VSCAYWSKP--AGAATSFLSLMSGAALPVPDAARAVELVSAPSRGDSSTA
. * * .: : :.
Mb3480c|M.bovis_AF2122/97 DAVYLPPGRSAYVAARSLSGGGT--GTRYLVTDTGVRFAIHDDDVAH---
Rv3450c|M.tuberculosis_H37Rv DAVYLPPGRSAYVAARSLSGGGT--GTRYLVTDTGVRFAIHDDDVAH---
MMAR_1099|M.marinum_M DAVYLPPGRSAYVTSRGLSGSGAPLGRRYLVTDAGVRYLIRDEDTAR---
MUL_0857|M.ulcerans_Agy99 DAVYLPPGRSAYVTSRGLSGSGAPLGRRYLVTDAGVRYLIHDEDTAR---
MAV_4392|M.avium_104 DAVYLPPGRSAYVRA------GANAGTRYLITDTGVRFAVPDDDAAA---
Mflv_4962|M.gilvum_PYR-GCK -EVAVPPGRSLYVRAVGLTGGDHSGGLLFLVTDAAIMFGIRDADAAA---
Mvan_1444|M.vanbaalenii_PYR-1 DAVSVPAGRSVYVRSVSLTGAGHGTGSLFLVTDAGVLFGVRDDESAA---
TH_4296|M.thermoresistible__bu DAVGMPGGRYAFVRATGLTGDGGIAGSLFLLTDSGVVFGVHDEEAAQ---
MSMEG_1534|M.smegmatis_MC2_155 DNIGVPAGRSVLVEATGLAGG---RGPLYLLTDHGVLHGIQDADAAR---
MAB_3759c|M.abscessus_ATCC_199 DTAYIPPARSAFVRSTGLGGDTGRAESLYLILDTGVRFGIGNLEAAK---
MLBr_02536|M.leprae_Br4923 SRVVLTPGTGYFAQTVGVGSAAPATASLFWVSDTGVRYGIDTEADARSEA
:. . . : : : * .: . : *
Mb3480c|M.bovis_AF2122/97 ---------DLGLPTAAIPAPWPVLATLPSGPELSRANASVARDTVAPGP
Rv3450c|M.tuberculosis_H37Rv ---------DLGLPTAAIPAPWPVLATLPSGPELSRANASVARDTVAPGP
MMAR_1099|M.marinum_M ---------HLGLLTP-TAAPWPMLATLPEGPELSGANASVAYDVVAVDP
MUL_0857|M.ulcerans_Agy99 ---------HLGLLTP-TAAPWPMLATLPEGPELSGASASVAYDVVAVDP
MAV_4392|M.avium_104 ---------DLGLGQP-SPAPWPLLTLLPAGPELSRPNASVARDTVAGAP
Mflv_4962|M.gilvum_PYR-GCK ---------ALGLVGAGVPAPWPVLATVPRGPELSRDAATGR--------
Mvan_1444|M.vanbaalenii_PYR-1 ---------ALGLTAPAVPAPWPVLATLPRGPELGRDDASVLRDGTGAP-
TH_4296|M.thermoresistible__bu ---------WLGLPADARPAPWPMLARLPRGPELGAQAASVSRDGAPPA-
MSMEG_1534|M.smegmatis_MC2_155 ---------HLGLTGPAVPAPWAMLAMLPRGPELSSAAASVARDALPSTS
MAB_3759c|M.abscessus_ATCC_199 ---------ALGLNVEPVPAPWPIVGLLPPGPRLSKEDALVVHDGMPPDK
MLBr_02536|M.leprae_Br4923 TAGPGKIVEALGLKLPAVPIPWSILSLFAVGPTLSRADALLEHDGLAPDT
*** . **.:: .. ** *. *
Mb3480c|M.bovis_AF2122/97 -------------------------
Rv3450c|M.tuberculosis_H37Rv -------------------------
MMAR_1099|M.marinum_M PVPAPGPATECPGGQCRQQPHQADR
MUL_0857|M.ulcerans_Agy99 PVPAPGPATECPGGQCPQQPHQADR
MAV_4392|M.avium_104 -------------------------
Mflv_4962|M.gilvum_PYR-GCK -------------------------
Mvan_1444|M.vanbaalenii_PYR-1 -------------------------
TH_4296|M.thermoresistible__bu -------------------------
MSMEG_1534|M.smegmatis_MC2_155 -------------------------
MAB_3759c|M.abscessus_ATCC_199 TGLAINPSTSATQAPS---------
MLBr_02536|M.leprae_Br4923 RAGRTTTAYGEHR------------