For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
PMGVAIEVKGLTKSFGSSRIWEDVTLDIPAGEVSVLLGPSGTGKSVFLKSLIGLLRPERGSILIDGTDII ECSAKELYEIRTLFGVLFQDGALFGSMNLYDNTAFPLREHTKKKESEIRDIVMEKLQLVGLGGDEKKFPG EISGGMRKRAGLARALVLDPQIILCDEPDSGLDPVRTAYLSQLIMDINAQIDATILIVTHNVNIARTVPD NMGMLFRKHLVMFGPREVLLTSDEPVVRQFLNGRRIGPIGMSEEKDESTMAEEAALLEAGHYAGGAEEVE GVPPQITVTPGMPKRKAVARRQARVRAMLPTLPKGAQAAILDDLEGAHNYQAHEFGD*
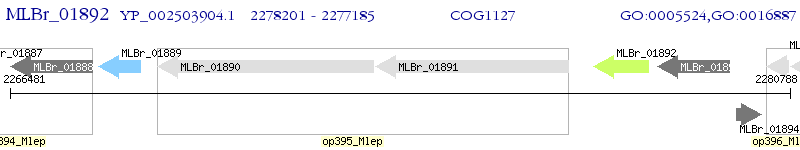
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. leprae Br4923 | MLBr_01892 | - | - | 100% (338) | ABC transporter ATP-binding protein |
| M. leprae Br4923 | MLBr_00669 | ftsE | 2e-27 | 33.33% (201) | putative cell division ATP-binding protein |
| M. leprae Br4923 | MLBr_01122 | - | 3e-23 | 30.14% (219) | putative ABC transport protein, ATP-binding component |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0674 | mkl | 1e-179 | 93.15% (336) | ribonucleotide ABC transporter ATP-binding protein |
| M. gilvum PYR-GCK | Mflv_5099 | - | 1e-162 | 83.88% (335) | ABC transporter related |
| M. tuberculosis H37Rv | Rv0655 | mkl | 1e-179 | 93.15% (336) | ribonucleotide ABC transporter ATP-binding protein |
| M. abscessus ATCC 19977 | MAB_3871c | - | 1e-160 | 81.55% (336) | ribonucleotide ABC transporter ATP-binding protein |
| M. marinum M | MMAR_0994 | mkl | 1e-172 | 92.66% (327) | ribonucleotide-transport ATP-binding protein ABC transporter, |
| M. avium 104 | MAV_4504 | - | 1e-172 | 88.69% (336) | ABC transporter, ATP-binding protein |
| M. smegmatis MC2 155 | MSMEG_1366 | - | 1e-157 | 84.26% (324) | ABC transporter, ATP-binding protein |
| M. thermoresistible (build 8) | TH_4213 | - | 1e-154 | 84.94% (312) | PUTATIVE ABC transporter-related protein |
| M. ulcerans Agy99 | MUL_0745 | mkl | 1e-172 | 92.35% (327) | ribonucleotide-transport ATP-binding protein ABC transporter, |
| M. vanbaalenii PYR-1 | Mvan_1254 | - | 1e-165 | 84.43% (334) | ABC transporter-related protein |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_0994|M.marinum_M -----------------------MGVAIEVNGLTKSFGSSRIWEDVTLTI
MUL_0745|M.ulcerans_Agy99 -----------------------MGVAIEVNGLTKSFGSSRIWEDVTLTI
Mb0674|M.bovis_AF2122/97 MRYSDSYHTTGRWQPRASTEGFPMGVSIEVNGLTKSFGSSRIWEDVTLTI
Rv0655|M.tuberculosis_H37Rv MRYSDSYHTTGRWQPRASTEGFPMGVSIEVNGLTKSFGSSRIWEDVTLTI
MLBr_01892|M.leprae_Br4923 ---------------------MPMGVAIEVKGLTKSFGSSRIWEDVTLDI
MAV_4504|M.avium_104 -----------------------MGVAIEVNGLTKSFGSSRIWEDVTLEI
MSMEG_1366|M.smegmatis_MC2_155 -----------------------MGVQIDVTGLSKSFGSSKIWEDVTMSI
MAB_3871c|M.abscessus_ATCC_199 ----------------MLRKERLVGAEVSVEGLTKSFGSQRIWENVTLSL
TH_4213|M.thermoresistible__bu -----------------------------------------------MDI
Mvan_1254|M.vanbaalenii_PYR-1 -----------------------MGIGIQIEGLTKSFGAQRIWEDVTFDL
Mflv_5099|M.gilvum_PYR-GCK -----------------------MGIGIQVEGLTKSFGSQRIWEDVTFDL
: :
MMAR_0994|M.marinum_M PAGEVSVLLGPSGTGKSVFLKSLIGLLRPERGSILIDGTDIIECSAKELY
MUL_0745|M.ulcerans_Agy99 PAGEVSVLLGPSGTGKSVFLKSLIGLLRPERGSILIDGTDIIECSAKELY
Mb0674|M.bovis_AF2122/97 PAGEVSVLLGPSGTGKSVFLKSLIGLLRPERGSIIIDGTDIIECSAKELY
Rv0655|M.tuberculosis_H37Rv PAGEVSVLLGPSGTGKSVFLKSLIGLLRPERGSIIIDGTDIIECSAKELY
MLBr_01892|M.leprae_Br4923 PAGEVSVLLGPSGTGKSVFLKSLIGLLRPERGSILIDGTDIIECSAKELY
MAV_4504|M.avium_104 PAGEVSVLLGPSGTGKSVFLKSLIGLLRPERGSVVIDGTDILQCSAKELY
MSMEG_1366|M.smegmatis_MC2_155 PAGEVSVLLGPSGTGKSVFLKSLIGLLRPERGSIVIDGTDILQCSAKELY
MAB_3871c|M.abscessus_ATCC_199 PPGEVSVLLGPSGTGKSVFLKSLIGLLRPEQGAIYIDGTNIMECSSKELY
TH_4213|M.thermoresistible__bu PAGEVSVLLGPSGTGKSVFLKSLIGLLRPERGKIIVDGTNIIDCTAKELY
Mvan_1254|M.vanbaalenii_PYR-1 PAGEVSVLLGPSGTGKSVFLKSLIGLLRPERGKIIVDGTNIIECSAKELY
Mflv_5099|M.gilvum_PYR-GCK PAGEVSVLLGPSGTGKSVFLKSLIGLLRPERGKIVVDGTNIIECSAKELY
*.****************************:* : :***:*::*::****
MMAR_0994|M.marinum_M EIRTLFGVLFQDGALFGSMNLYDNTAFPLREHTKKKESEIRDIVMEKLAM
MUL_0745|M.ulcerans_Agy99 EIRTLFGVLFQDGALFGSMNLYDNTAFPLREHTKKKESEIRDIVMEKLAM
Mb0674|M.bovis_AF2122/97 EIRTLFGVLFQDGALFGSMNLYDNTAFPLREHTKKKESEIRDIVMEKLAL
Rv0655|M.tuberculosis_H37Rv EIRTLFGVLFQDGALFGSMNLYDNTAFPLREHTKKKESEIRDIVMEKLAL
MLBr_01892|M.leprae_Br4923 EIRTLFGVLFQDGALFGSMNLYDNTAFPLREHTKKKESEIRDIVMEKLQL
MAV_4504|M.avium_104 EIRTLFGVMFQDGALFGSMNLFDNAAFPLREHTKKKESEIRDIVMEKLEL
MSMEG_1366|M.smegmatis_MC2_155 EIRTLFGVLFQDGALFGSMNIYDNTAFPLREHTKKSESEIRKIVMEKLDL
MAB_3871c|M.abscessus_ATCC_199 EIRKLFGVMFQDGALFGSMNLYDNTAFPLREHTKKKESEIRDIVMQKLDV
TH_4213|M.thermoresistible__bu EIRTLFGVMFQDGALFGSMSIYDNTAFPLREHTKKKESEIRKIVMEKLEL
Mvan_1254|M.vanbaalenii_PYR-1 EIRTLFGVMFQDGALFGSMSLYDNTAFPLREHTKKKESEIRQIVMEKLDL
Mflv_5099|M.gilvum_PYR-GCK EIRTLFGVMFQDGALFGSMSLYDNTAFPLREHTKKKESEIRKIVMEKLDL
***.****:**********.::**:**********.*****.***:** :
MMAR_0994|M.marinum_M VGLTGDEKKFPGEISGGMRKRAGLARALVLDPQIILCDEPDSGLDPVRTA
MUL_0745|M.ulcerans_Agy99 VGLTGDEKKFPGEISGGMRKRAGLARALVLDPQIILCDEPDSGLDPVRTA
Mb0674|M.bovis_AF2122/97 VGLGGDEKKFPGEISGGMRKRAGLARALVLDPQIILCDEPDSGLDPVRTA
Rv0655|M.tuberculosis_H37Rv VGLGGDEKKFPGEISGGMRKRAGLARALVLDPQIILCDEPDSGLDPVRTA
MLBr_01892|M.leprae_Br4923 VGLGGDEKKFPGEISGGMRKRAGLARALVLDPQIILCDEPDSGLDPVRTA
MAV_4504|M.avium_104 VGLGGDEKKFPGEISGGMRKRASLARALVMDPQIILCDEPDSGLDPVRTA
MSMEG_1366|M.smegmatis_MC2_155 VGMPNDGHKFPGEISGGMRKRAGLARALVLDPEIILCDEPDSGLDPVRTA
MAB_3871c|M.abscessus_ATCC_199 VGLTGHETKFPGEISGGMRKRAGLARSLVLDPQIILCDEPDSGLDPVRTA
TH_4213|M.thermoresistible__bu VGLAGDENKFPGEISGGMRKRAGLARSLVLDPQIILCDEPDSGLDPVRTA
Mvan_1254|M.vanbaalenii_PYR-1 VGLGGDESKFPGEISGGMRKRAGLARSLVLDPQIILCDEPDSGLDPVRTA
Mflv_5099|M.gilvum_PYR-GCK VGLGGDEDKFPGEISGGMRKRAGLARSLVLDPQIILCDEPDSGLDPVRTA
**: .. **************.***:**:**:*****************
MMAR_0994|M.marinum_M YLSQLIMDINAQIDATILIVTHNINIARTVPDNMGMLFRKHLVMFGPREV
MUL_0745|M.ulcerans_Agy99 YLSQLIMDINAQIDATILIVTHNINIARTVPDNMGMLFRKHLVMFGPREV
Mb0674|M.bovis_AF2122/97 YLSQLIMDINAQIDATILIVTHNINIARTVPDNMGMLFRKHLVMFGPREV
Rv0655|M.tuberculosis_H37Rv YLSQLIMDINAQIDATILIVTHNINIARTVPDNMGMLFRKHLVMFGPREV
MLBr_01892|M.leprae_Br4923 YLSQLIMDINAQIDATILIVTHNVNIARTVPDNMGMLFRKHLVMFGPREV
MAV_4504|M.avium_104 YLSQLILDINAQIDATVLIVTHNINIARTVPDNMGMLFRRKLVMFGPREV
MSMEG_1366|M.smegmatis_MC2_155 YLSQLLIDINAQIDATVLIVTHNINIARTVPDNMGMLFRKQLVMFGPREV
MAB_3871c|M.abscessus_ATCC_199 YLSQLLIDINAQIDATILIVTHNINIARTVPDNMGMLYRRNLVMFGPREV
TH_4213|M.thermoresistible__bu YLSQLLIDINAQIDCTILIVTHNINIARTVPDNIGMLFRKRLVMFGPREV
Mvan_1254|M.vanbaalenii_PYR-1 YLSQLLIDINAQIDCTILIVTHNINIARTVPDNMGMLFRKKLVMFGPREV
Mflv_5099|M.gilvum_PYR-GCK YLSQLLIDINAQIDCTILIVTHNINIARTVPDNMGMLFRKHLVMFGPREV
*****::*******.*:******:*********:***:*:.*********
MMAR_0994|M.marinum_M LLTSDEPVVRQFLNGRRIGPIGMSEEKDEATMAEEQAMIDAGQHAGGTED
MUL_0745|M.ulcerans_Agy99 LLASDEPVVRQFLNGRRIGPIGMSEEKDEATMAEEQAMIDAGQHAGGTED
Mb0674|M.bovis_AF2122/97 LLTSDEPVVRQFLNGRRIGPIGMSEEKDEATMAEEQALLDAGHHAGGVEE
Rv0655|M.tuberculosis_H37Rv LLTSDEPVVRQFLNGRRIGPIGMSEEKDEATMAEEQALLDAGHHAGGVEE
MLBr_01892|M.leprae_Br4923 LLTSDEPVVRQFLNGRRIGPIGMSEEKDESTMAEEAALLEAGHYAGGAEE
MAV_4504|M.avium_104 LLTSDEPVVKQFLNGRRIGPIGMSEEKDEATMAEEQAALEAGQHAGGVEE
MSMEG_1366|M.smegmatis_MC2_155 LLTSEEPVVKQFLNGRRIGPIGMSEEKDEATAAAEQALVDAGQHDGGVEE
MAB_3871c|M.abscessus_ATCC_199 LLTSDEPVIKQFLNGRRIGPIGMSEEKDEATMAEEQAHLDAGHHDGGVEE
TH_4213|M.thermoresistible__bu LLTSDEPVVRQFLNGRRIGPIGMSEEKDELTMAEEQAMVDAGHHDGGVEE
Mvan_1254|M.vanbaalenii_PYR-1 LLTSDEPVVKQFLNGRRLGPIGMSEEKDESTMAEEQAMADAGHHDGGVEE
Mflv_5099|M.gilvum_PYR-GCK LLTSDEPVVKQFLNGRRIGPIGMSEEKDESTMAEEQAMVDAGHHDGGTEE
**:*:***::*******:*********** * * * * :**:: **.*:
MMAR_0994|M.marinum_M IEGVPPQIQATPGMPERKAVARRQARVREMLHTLPKDAQAAILEDLEG--
MUL_0745|M.ulcerans_Agy99 IEGVPPQIQATPGMPERKAVARRQARVREMLHTLPKDAQAAILEDLEG--
Mb0674|M.bovis_AF2122/97 IEGVPPQISATPGMPERKAVARRQARVREMLHTLPKKAQAAILDDLEG--
Rv0655|M.tuberculosis_H37Rv IEGVPPQISATPGMPERKAVARRQARVREMLHTLPKKAQAAILDDLEG--
MLBr_01892|M.leprae_Br4923 VEGVPPQITVTPGMPKRKAVARRQARVRAMLPTLPKGAQAAILDDLEG--
MAV_4504|M.avium_104 IEGVPPQIVASPGMPERKAVARRQARVREILHTLPQKAQAAILDDLEG--
MSMEG_1366|M.smegmatis_MC2_155 IEGVPPQLQATPGMPERKAVARRKARVREILHTLPPAAQAAILEELDRDQ
MAB_3871c|M.abscessus_ATCC_199 IEGVPPQIQATPGMPERKAVGRRQARVREIMHQLPPNAQAAIQDSFAG--
TH_4213|M.thermoresistible__bu IVGVPPQLTATPGMPERQAVHRRRARVREILHTLPPAAQAAIRDDLEG--
Mvan_1254|M.vanbaalenii_PYR-1 IEGVPPQIQATPGMPERQAVARRQARVREIMHTLPPAAQAAIRDDLEG--
Mflv_5099|M.gilvum_PYR-GCK IEGVPPQVMATPGMPERKAVGRRQARVREIMHTLPPAAQEAIRADLEN--
: *****: .:****:*:** **:**** :: ** ** ** .:
MMAR_0994|M.marinum_M ---------------THR--------------------------------
MUL_0745|M.ulcerans_Agy99 ---------------THR--------------------------------
Mb0674|M.bovis_AF2122/97 ---------------THKYA--VHEIGQ----------------------
Rv0655|M.tuberculosis_H37Rv ---------------THKYA--VHEIGQ----------------------
MLBr_01892|M.leprae_Br4923 ---------------AHNYQ--AHEFGD----------------------
MAV_4504|M.avium_104 ---------------THKYR--AHEVGD----------------------
MSMEG_1366|M.smegmatis_MC2_155 PQLSAPTTQTAATAPVENYD--DSPTGVIEV--------PKQA-------
MAB_3871c|M.abscessus_ATCC_199 ---------------THQYR--VHDFGDEPTGVTTATLAPPQAEQPTQSI
TH_4213|M.thermoresistible__bu ---------------THKYR--ADEFGDSGAGQHRAHEDDAPTASIPVAG
Mvan_1254|M.vanbaalenii_PYR-1 ---------------THQFP--SHQFAGEPAVR---HEDDAPTGRIPVSG
Mflv_5099|M.gilvum_PYR-GCK ---------------TQQTQPYPDGYSDGTTAHHARHDDETPTGRIPVAG
...
MMAR_0994|M.marinum_M -----------------
MUL_0745|M.ulcerans_Agy99 -----------------
Mb0674|M.bovis_AF2122/97 -----------------
Rv0655|M.tuberculosis_H37Rv -----------------
MLBr_01892|M.leprae_Br4923 -----------------
MAV_4504|M.avium_104 -----------------
MSMEG_1366|M.smegmatis_MC2_155 -----------------
MAB_3871c|M.abscessus_ATCC_199 DTSDDDTPTGQIPPVRG
TH_4213|M.thermoresistible__bu EQ---------------
Mvan_1254|M.vanbaalenii_PYR-1 ER---------------
Mflv_5099|M.gilvum_PYR-GCK ER---------------