For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MCAHFNTSMTNTGDDTEQSDEVGSISRLRGRKTHRTRALTKNHPRSIALAYALLAPSLFGVVTFLLLPII VVIWLSLFRWDLLGPLHYVGLANWRSVLTDPNFANSLIVTAVFVAIVVPTQTMLGLLIATMLARQLPGTG VFRTLYVLPWICAPLAIAVLWRWLLAPTDGAFSMVLRHRIEWLSDPSLALPVVSAVVVWTNVGYVSLSFL AGLLSIPEDINAAARTDGADAWQRFWRITLPMLRPTMFFVLVTGIVSTAQVFDTVYALTDGGPKSRTDLV AHRIYAEAFGSAAIGRASVMAVLLFVVLFGVTVVQHLYFQRRISYELT
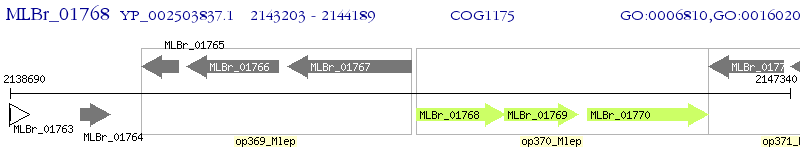
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. leprae Br4923 | MLBr_01768 | uspA | - | 100% (328) | sugar transport integral membrane protein |
| M. leprae Br4923 | MLBr_01426 | - | 1e-40 | 32.46% (305) | putative ABC-transport protein, inner membrane component |
| M. leprae Br4923 | MLBr_01087 | - | 6e-35 | 30.22% (268) | putative ABC-transport protein, inner membrane component |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2343 | uspA | 1e-138 | 83.80% (284) | sugar-transport integral membrane protein ABC transporter |
| M. gilvum PYR-GCK | Mflv_3852 | - | 3e-33 | 28.67% (279) | binding-protein-dependent transport systems inner membrane |
| M. tuberculosis H37Rv | Rv2316 | uspA | 1e-137 | 83.45% (284) | sugar-transport integral membrane protein ABC transporter |
| M. abscessus ATCC 19977 | MAB_1717c | - | 1e-117 | 68.90% (283) | sugar ABC transporter, permease protein |
| M. marinum M | MMAR_3617 | uspA | 1e-135 | 81.34% (284) | sugar-transport integral membrane protein ABC transporter |
| M. avium 104 | MAV_2090 | uspA | 1e-139 | 84.91% (285) | ABC transporter, permease protein UspA |
| M. smegmatis MC2 155 | MSMEG_4466 | - | 1e-132 | 80.28% (284) | ABC transporter, permease protein UspA |
| M. thermoresistible (build 8) | TH_4307 | - | 3e-35 | 31.75% (274) | PUTATIVE binding-protein-dependent transport systems inner |
| M. ulcerans Agy99 | MUL_1248 | uspA | 1e-135 | 81.34% (284) | sugar-transport integral membrane protein ABC transporter |
| M. vanbaalenii PYR-1 | Mvan_2543 | - | 2e-34 | 28.28% (297) | binding-protein-dependent transport systems inner membrane |
CLUSTAL 2.0.9 multiple sequence alignment
Mb2343|M.bovis_AF2122/97 --------------------------------------MRDAPRRRTALA
Rv2316|M.tuberculosis_H37Rv --------------------------------------MRDAPRRRTALA
MLBr_01768|M.leprae_Br4923 MCAHFNTSMTNTGDDTEQSDEVGSISRLRGRKTHRTRALTKNHPRSIALA
MAV_2090|M.avium_104 ----------------------------------MASAERRVAPRSTALG
MMAR_3617|M.marinum_M --------------------------------------MRATPRRTTALA
MUL_1248|M.ulcerans_Agy99 --------------------------------------MRATPRRTTALA
MSMEG_4466|M.smegmatis_MC2_155 -----------------------MAPWRLLTANRFNEPMATPRVRTTALA
MAB_1717c|M.abscessus_ATCC_199 ----------------------------------------MSRSRQTAVA
Mflv_3852|M.gilvum_PYR-GCK ---------------------MTAPADAPIQKDPGKPSVSERAKGERRLG
Mvan_2543|M.vanbaalenii_PYR-1 ---------------------MTALHEAPIQKDPGTPAVSERARGERRLG
TH_4307|M.thermoresistible__bu ---------------------MTAAAEKADPTPAAAVARTDDTRSERRLA
:.
Mb2343|M.bovis_AF2122/97 YALLAPSLVGVVAFLLLPILVVVWLSLHRWDLLGP--LRYVGLTNWRSVL
Rv2316|M.tuberculosis_H37Rv YALLAPSLVGVVAFLLLPILVVVWLSLHRWDLLGP--LRYVGLTNWRSVL
MLBr_01768|M.leprae_Br4923 YALLAPSLFGVVTFLLLPIIVVIWLSLFRWDLLGP--LHYVGLANWRSVL
MAV_2090|M.avium_104 YALLAPSLFGVLAFLLLPILVVIWLSLCRWDLLGP--LRFVGLSNWRSVL
MMAR_3617|M.marinum_M YALLAPSLFGVLAFLLLPILVVIWLSLYRWDLLGP--LRYVGLANWRSVL
MUL_1248|M.ulcerans_Agy99 YALLAPSLFGVLAFLLLPILVVIWLSLYRWDLLGP--LRYVGLANWRSVL
MSMEG_4466|M.smegmatis_MC2_155 YALVAPSLFGVVTFLLLPMLVVVWLSLHRWDLLGP--IEFVGLDNWTSVL
MAB_1717c|M.abscessus_ATCC_199 YGLLAPSLFGVAAFLLLPILVVVWLSLCRWDLLGP--IEFVGLDNWRSVL
Mflv_3852|M.gilvum_PYR-GCK LYLTLPSYVVMLLVTAYPLGYALVLSLYNFRLTDPEGRSFVGLGNYLVIL
Mvan_2543|M.vanbaalenii_PYR-1 IYLTLPSYVVMLLVTAYPLGYALVLSLYNFRLTDPEGRSFVGLSNYLVIL
TH_4307|M.thermoresistible__bu YWLIAPAVLLMVAVTGYPIAYAVWLSLLRYNLAQPDDVSFVGLENYVTVL
* *: . : . *: .: *** .: * * :*** *: :*
Mb2343|M.bovis_AF2122/97 TDSGFADSLVVTAVFVAIVVPAQTVLGLLAASLLARRLPGTGLFRTLYVL
Rv2316|M.tuberculosis_H37Rv TDSGFADSLVVTAVFVAIVVPAQTVLGLLAASLLARRLPGTGLFRTLYVL
MLBr_01768|M.leprae_Br4923 TDPNFANSLIVTAVFVAIVVPTQTMLGLLIATMLARQLPGTGVFRTLYVL
MAV_2090|M.avium_104 TDAGFGNSLMVTAVFVAMVVPAQTALGLLAATMLARRLPGTGLFRTVYVL
MMAR_3617|M.marinum_M RDGDFGNSLVVTAIFVAIVVPAQTVLGLLAAMMLTRRLPGTNFFRTLYVL
MUL_1248|M.ulcerans_Agy99 RDGDFGNSLVVTAIFVAIVVPAQTVLGLLAAMMLTRRLPGTNFFRTLYVL
MSMEG_4466|M.smegmatis_MC2_155 TDPAFGKSLVVTLLFMALVVPTQTVLGLVAATMLARGLPGTGFFRTVYVL
MAB_1717c|M.abscessus_ATCC_199 TDGTFGHSLLVTLLFVALVIPVQTALGLAAATLLVRGLPGTVLFRTIYVI
Mflv_3852|M.gilvum_PYR-GCK TDPIWWNDFVTTLAITVVTVAIELVLGFWFAFVMLRIVRGRGPLRTAILI
Mvan_2543|M.vanbaalenii_PYR-1 TDPLWWNDFVTTLAITVVTVAIELVLGFWFAFVMLRIVRGRGPLRTAILI
TH_4307|M.thermoresistible__bu TDEYWWTAFAVTLGITVVSVAIEFVLGLALALVMHRTIFGKGVVRTAILI
* : : .* : .: :. : **: * :: * : * .** ::
Mb2343|M.bovis_AF2122/97 PWICAPLAIAVMWRWILAPTDGAISTVLG-HRIEWLTDPGLALPVVSAVV
Rv2316|M.tuberculosis_H37Rv PWICAPLAIAVMWRWIVAPTDGAISTVLG-HRIEWLTDPGLALPVVSAVV
MLBr_01768|M.leprae_Br4923 PWICAPLAIAVLWRWLLAPTDGAFSMVLR-HRIEWLSDPSLALPVVSAVV
MAV_2090|M.avium_104 PWICAPLAIAVLWRWILAPTDGAVSAVLG-HSIEWLSDPSFALPLVSAVV
MMAR_3617|M.marinum_M PWICAPLAIAVMWRWILAPTDGAISTLLG-RRIEWLTDPDLALPVVAAVV
MUL_1248|M.ulcerans_Agy99 PWICAPLAIAVMWRWILAPTDGAISTLLG-RRIEWLTDPDLALPVVAAVV
MSMEG_4466|M.smegmatis_MC2_155 PWICAPLAIAVLWRWILAPTDGALSTVLG-TRIEWLTDPSLALPVVSAVV
MAB_1717c|M.abscessus_ATCC_199 PWICAPLAIGVLWHWMLAPTDGAVNALLG-QRVEWLTDPSLALPVVAAVS
Mflv_3852|M.gilvum_PYR-GCK PYGIVTVVSAFIWRYAFAIDSGFVNQWLNLTDFDWFGDRWSAIFVICLSE
Mvan_2543|M.vanbaalenii_PYR-1 PYGIVTVVSAFIWRYAFAIDSGFVNQWLNLTEFDWFGDRWSAIFVICLSE
TH_4307|M.thermoresistible__bu PYGIVTVAASYSWYYAWTPGTGYLANLLPDGSAPLT-QQLPSLAIVVLAE
*: ..:. . * : : * . * : :: ::
Mb2343|M.bovis_AF2122/97 VWTNVGYVSLFFLAGLMAIPQDIHNAARTDGASAWQRFWRITLPMLRPTM
Rv2316|M.tuberculosis_H37Rv VWTNVGYVSLFFLAGLMAIPQDIHNAARTDGASAWQRFWRITLPMLRPTM
MLBr_01768|M.leprae_Br4923 VWTNVGYVSLSFLAGLLSIPEDINAAARTDGADAWQRFWRITLPMLRPTM
MAV_2090|M.avium_104 VWTNVGYVSLSFLAGLLAIPEDIHAAARTDGANAWQRFWRITMPMLRPTT
MMAR_3617|M.marinum_M IWTNVGYVSLSFLAGLLAIPDDIHAAARTDGANAWQRFWRITLPMLRPTT
MUL_1248|M.ulcerans_Agy99 IWTNVGYVSLSFLAGLLAIPDDIHAAARTDGANAWQRFWRITLPMLRPTT
MSMEG_4466|M.smegmatis_MC2_155 VWTNVGYVTLFFLAGILNIPADLHNAARTDGADAWQRFRHITLPMLRPTM
MAB_1717c|M.abscessus_ATCC_199 VWTNVGYVALFFMAGLLAIPGDVHNAARVDGATAWQRFRRITLPMLRPTF
Mflv_3852|M.gilvum_PYR-GCK IWKTTPFISLLLLAGLVQVPEDMQEAAKVDGATAWQRLWKITLPNMKAAI
Mvan_2543|M.vanbaalenii_PYR-1 IWKTTPFISLLLLAGLVQVPEDMQEAAKVDGATAWQRLWKITLPNMKAAI
TH_4307|M.thermoresistible__bu VWKTTPFMALLLLAGLALVPQELLNAAQMDGANAWQRLVKIILPMIKPAI
:*... :::* ::**: :* :: **: *** ****: :* :* ::.:
Mb2343|M.bovis_AF2122/97 FFVLVTGIISAAQVFDTVYALTGGGPQGSTDLVAHRIYAEAFGAAAIGRA
Rv2316|M.tuberculosis_H37Rv FFVLVTGIISAAQVFDTVYALTGGGPQGSTDLVAHRIYAEAFGAAAIGRA
MLBr_01768|M.leprae_Br4923 FFVLVTGIVSTAQVFDTVYALTDGGPKSRTDLVAHRIYAEAFGSAAIGRA
MAV_2090|M.avium_104 FFVLVTGIVSSAQVFDTVYALTGGGPAGSTDLVAHRIYAEAFGSAAIGRA
MMAR_3617|M.marinum_M FFVLVTGIVSAAQIFDIVYALTGGGPEGSTELVAHQIYAEAFGSAAIGRA
MUL_1248|M.ulcerans_Agy99 FFVLVTGIVSAAQIFDIVYALTGGGPEGSTELVAHQIYAEAFGSAAIGRA
MSMEG_4466|M.smegmatis_MC2_155 FFVLVTGIVSAAQVFDTVYALTAGGPQGRTDLIAHRIYAEAFGAAAVGRA
MAB_1717c|M.abscessus_ATCC_199 FFVLVTGVVSAVQTFDTVYALTSGGPQNRTDLAAHRVYAEAFEAAHVGRS
Mflv_3852|M.gilvum_PYR-GCK MVALLFRTLDAWRIFDNPYVMTAG--ANNTETISFLAYRQNVTLVNLGMG
Mvan_2543|M.vanbaalenii_PYR-1 MVALLFRTLDAWRIFDNPYVMTAG--ANNTETISFLAYRQNVTLVNLGMG
TH_4307|M.thermoresistible__bu LVALLFRTLDAFRIFDNIYILTGG--ANNTGSVSILGYNNLFKAFNLGLG
:..*: :.: : ** * :* * . * : * : . :* .
Mb2343|M.bovis_AF2122/97 SVMAVVLFVILVGATVVQHLYFRRRISYELT---
Rv2316|M.tuberculosis_H37Rv SVMAVVLFVILVGATVVQHLYFRRRISYELT---
MLBr_01768|M.leprae_Br4923 SVMAVLLFVVLFGVTVVQHLYFQRRISYELT---
MAV_2090|M.avium_104 SVMAVVLFVILIGVTLVQHLYFRRRISYDLT---
MMAR_3617|M.marinum_M SVMAVVLFVILIGVTFVQHMYFRRRISYDLT---
MUL_1248|M.ulcerans_Agy99 SVMAVVLFVILIGVTFVQHMYFRRRISYDLT---
MSMEG_4466|M.smegmatis_MC2_155 AVMALVLFVLLVGVTIVQHLYFRRRISYELT---
MAB_1717c|M.abscessus_ATCC_199 AAMSLILFLILVTITVIQHRYFRARVSYDLV---
Mflv_3852|M.gilvum_PYR-GCK SAVSVLLFLSVVAIAWIFIKVFKTDLSQVRGDQ-
Mvan_2543|M.vanbaalenii_PYR-1 SAVSVLLFLSVVGIAWIFIKVFKTDLSQVRGDQ-
TH_4307|M.thermoresistible__bu SAISVLIFLCVAVIAFIYIKIFGTSAPGSDAERS
:.:::::*: : : : * .