For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VTSNSDAGSAVDAGGPPRTVIIAAVVLTAATIGTILVLAATLHEPPQPVVITAVPAPQATTAACRSLTQA LPQRLGDYERAPVAQPAPDAVAAWRTGSDTEPVVLRCGLDGPAEFVVGSPIQAVDQVQWFEVDAKPKPAI DAGRSTWYTVDRPVYVALTLPSGSGPTPIQELSDVIDRTLAAIPISPAQSH
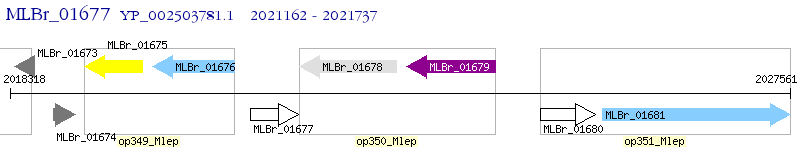
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. leprae Br4923 | MLBr_01677 | - | - | 100% (191) | putative secreted protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3004 | - | 7e-64 | 66.67% (177) | hypothetical protein Mb3004 |
| M. gilvum PYR-GCK | Mflv_4218 | - | 7e-46 | 47.92% (192) | hypothetical protein Mflv_4218 |
| M. tuberculosis H37Rv | Rv2980 | - | 7e-64 | 66.67% (177) | hypothetical protein Rv2980 |
| M. abscessus ATCC 19977 | MAB_3285 | - | 1e-31 | 38.42% (177) | hypothetical protein MAB_3285 |
| M. marinum M | MMAR_1734 | - | 2e-64 | 61.26% (191) | hypothetical protein MMAR_1734 |
| M. avium 104 | MAV_3831 | - | 2e-69 | 70.69% (174) | hypothetical protein MAV_3831 |
| M. smegmatis MC2 155 | MSMEG_2396 | - | 2e-50 | 53.26% (184) | hypothetical protein MSMEG_2396 |
| M. thermoresistible (build 8) | TH_2050 | - | 3e-48 | 54.29% (175) | POSSIBLE CONSERVED SECRETED PROTEIN |
| M. ulcerans Agy99 | MUL_1977 | - | 4e-64 | 61.26% (191) | hypothetical protein MUL_1977 |
| M. vanbaalenii PYR-1 | Mvan_2149 | - | 2e-44 | 46.31% (203) | hypothetical protein Mvan_2149 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_4218|M.gilvum_PYR-GCK --------------MDSET-------ADGPPRAVLIAAAAV-AVGAVVAL
Mvan_2149|M.vanbaalenii_PYR-1 --------------MSSES-------TDGPPRAALIASLIV-AVGGLVAV
MLBr_01677|M.leprae_Br4923 --------------MTSNSDAGSAVDAGGPPRTVIIAAVVL-TAATIGTI
MAV_3831|M.avium_104 --------------MATESEAD-----GGPPRAVIMVAVAL-AVTTIGVI
Mb3004|M.bovis_AF2122/97 --------------MTGES--------DGPPRAVLIAAAAL-AAAVIGVI
Rv2980|M.tuberculosis_H37Rv --------------MTGES--------DGPPRAVLIAAAAL-AAAVIGVI
MMAR_1734|M.marinum_M MGQESRVEPEVSDAVTGDTPASPQPDPDGPPRVLIIAAALL-AVAALGTI
MUL_1977|M.ulcerans_Agy99 --------------MTGDTPANPQPDPDGPPRVLIIAAALL-AVAALGTI
MSMEG_2396|M.smegmatis_MC2_155 --------------------MTDPHTTDGPPRALLIAAVVV-AVSVLITV
TH_2050|M.thermoresistible__bu ----------VADKSHGESHEIAPEEPDAPPRALLIAAIAV-AVVAVVAI
MAB_3285|M.abscessus_ATCC_1997 --------------MQSED---------GPPRKLLIGIAVFGAIAALAVL
.*** :: . : : .:
Mflv_4218|M.gilvum_PYR-GCK LIVVAVMQREPAQKPVAVSGIPAPQADSPQCRSLVEALP-EQLGDYRRAQ
Mvan_2149|M.vanbaalenii_PYR-1 LVVAAVLQRTPEPQPVAIAGIPAPRAESPQCRALVDALP-DQLGDYRRAA
MLBr_01677|M.leprae_Br4923 LVLAATLHEPP--QPVVITAVPAPQATTAACRSLTQALP-QRLGDYERAP
MAV_3831|M.avium_104 LAIAATREAPP--QPVAVAAFPAPHAGDAACRALLDALP-QRLGDYQRAP
Mb3004|M.bovis_AF2122/97 LVVAANRQPPE--RPVVIPAVPAPQATGPGCKALLAALP-QRLGEYRRAP
Rv2980|M.tuberculosis_H37Rv LVVAANRQPPE--RPVVIPAVPAPQATGPGCKALLAALP-QRLGEYRRAP
MMAR_1734|M.marinum_M LVIAANREVPR--QPVAVPSVPAPQAASPACHALLTALP-QRLGDYQRAP
MUL_1977|M.ulcerans_Agy99 LVIAANREVPR--QPVAVPSVPAPQAASPACHALLTALP-QRLGDYQRAP
MSMEG_2396|M.smegmatis_MC2_155 LGIAASHKQSPEQEPVAISSVPAPQADSPECHALMDGLP-EQLGDFRRAP
TH_2050|M.thermoresistible__bu LAVAASRQGDPARQPVAIAAVPAPHADDPGCQELLDALP-EQLGDFERAP
MAB_3285|M.abscessus_ATCC_1997 FGVLKYAQFQNETRPLSVANIPAPQANSPLCVDMASAYPGKMAGDWSRVE
: : . .*: :. .***:* . * : . * . *:: *.
Mflv_4218|M.gilvum_PYR-GCK LVEPAPQGAAAWQPEAPDGEALILRCGLDRPAEFVTGAPLQVVDAVSWFR
Mvan_2149|M.vanbaalenii_PYR-1 LLEPAPPGTAAWQREEPGSEALILRCGLDRPAEFVAGAPVQVVDAVEWFR
MLBr_01677|M.leprae_Br4923 VAQPAPDAVAAWRT-GSDTEPVVLRCGLDGPAEFVVGSPIQAVDQVQWFE
MAV_3831|M.avium_104 LEQPAPAGAAAWRT-GPDSEPVVLRCGLDRPAEFVAGSPIQVVDRVQWFA
Mb3004|M.bovis_AF2122/97 VAEPTTAGATAWRT-GPNSTPVILRCGLDRPAEFVVGSAIQVVDRVQWFQ
Rv2980|M.tuberculosis_H37Rv VAEPTTAGATAWRT-GPNSTPVILRCGLDRPAEFVVGSAIQVVDRVQWFQ
MMAR_1734|M.marinum_M MVQPAPQGTGAWSS-AAADEPVILRCGLDRPADFVVGSPIQVVDRVQWFR
MUL_1977|M.ulcerans_Agy99 MVQPAPQGTGAWSS-AAADEPVILRCGLDRPADFVVGSPIQVVDRVQWFR
MSMEG_2396|M.smegmatis_MC2_155 TVEPTPQGTAAWRG-AADMEPIILRCGLERPVDFVVGAPLQVVDLVQWFR
TH_2050|M.thermoresistible__bu TVDPAPAGTAAWRR-GDD--TVILRCGLDRPADFVVGAPLQMVDDVQWFR
MAB_3285|M.abscessus_ATCC_1997 IAEPKPPAAAAWRR---GQDSVIARCGVERPAEFAVGTSVEQVNGVQWFR
:* . .. ** .:: ***:: *.:*..*:.:: *: *.**
Mflv_4218|M.gilvum_PYR-GCK VGEAGAGNDRVGEAGAG-----ESGVAD--GDRSTWFAVDRPVYVALTLP
Mvan_2149|M.vanbaalenii_PYR-1 VGEAGAGNDRVGEAGAGNDRVGEAGADDPAGERSTWFAVDRPVYVALTIP
MLBr_01677|M.leprae_Br4923 VDAKP-----------------KPAIDA---GRSTWYTVDRPVYVALTLP
MAV_3831|M.avium_104 VRPDP-----------------QSAGDA---GRSTWYTVDRPVYVALTLP
Mb3004|M.bovis_AF2122/97 VAAQ-------------------NPDEP---GRSTWYTVDRPVYVALTLP
Rv2980|M.tuberculosis_H37Rv VAAQ-------------------NPDEP---GRSTWYTVDRPVYVALTLP
MMAR_1734|M.marinum_M VSDQT-----------------EAAGTATAGSRSTWYTVDRPVYVALTLP
MUL_1977|M.ulcerans_Agy99 VSDQT-----------------EAAGTATAGSRSTWYTVDRPVYVALTLP
MSMEG_2396|M.smegmatis_MC2_155 VGDTGAAQP------------GDADAGAAEDTRSTWYAVDRPVYVALTLP
TH_2050|M.thermoresistible__bu V---------------------------AEADRTTWYAVDRPLYVALTLP
MAB_3285|M.abscessus_ATCC_1997 VSDSA-------------------------LASTTWFAVDRGVYVAVTVP
* :**::*** :***:*:*
Mflv_4218|M.gilvum_PYR-GCK RDSGPTAIQTLSTIIAETLPAQTPQPAPLG-
Mvan_2149|M.vanbaalenii_PYR-1 ADSGPTPIQTLSKIIADTLQSRPPDPAPIG-
MLBr_01677|M.leprae_Br4923 SGSGPTPIQELSDVIDRTLAAIPISPAQSH-
MAV_3831|M.avium_104 SGSGPAPIQRLSDVIEQRIAAAPVNPGPPAG
Mb3004|M.bovis_AF2122/97 SGSGPTAIQELSDVIDHTIPAVPIDPAPAR-
Rv2980|M.tuberculosis_H37Rv SGSGPTAIQELSDVIDHTIPAVPIDPAPAR-
MMAR_1734|M.marinum_M AGSGPTPIQELSEIIDRAIAPVPIDPAPAG-
MUL_1977|M.ulcerans_Agy99 AGSGPTPIQELSEIIDRAIAPAPIDPAPAG-
MSMEG_2396|M.smegmatis_MC2_155 SGSGPTPIQLISGQIARAMPEKPVEPGPAR-
TH_2050|M.thermoresistible__bu ADSGPSPIQGLSQTIAKVLPAKEIDPAPAR-
MAB_3285|M.abscessus_ATCC_1997 DGAGSEPLVEMSDAIAKALPAVKPDPKPLPR
.:*. .: :* * : .*