For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TESWESQVAGSAVPKPQLMSANGVSAAAAGRGERLNHAGNPGHRSKYSRVLLKLGGEMFGGGQVGLDLDV VAQVARQIAEVVRGGVQVAVVIGGGNFFRGAQLQQRGMERTRSDYMGMLGTVMNSLALQDFLEKEGIATR VQTAITMGQVAEPYLPLRAVRHLEKGRVVIFGAGMGLPYFSTDTTAAQRALEIGADVVLMAKAVDGVFVE DPRVNPEAELLTAISHREVIDRGLRVADATAFSLCMDNGMPILVFNLLTNGNIARAVAGEKIGTLVTT*
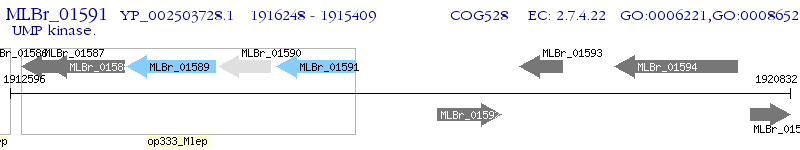
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. leprae Br4923 | MLBr_01591 | pyrH | - | 100% (279) | uridylate kinase |
| M. leprae Br4923 | MLBr_02323 | ask | 4e-07 | 25.63% (199) | aspartate kinase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2907c | pyrH | 1e-124 | 85.04% (274) | uridylate kinase |
| M. gilvum PYR-GCK | Mflv_4124 | pyrH | 1e-119 | 90.34% (238) | uridylate kinase |
| M. tuberculosis H37Rv | Rv2883c | pyrH | 1e-124 | 85.04% (274) | uridylate kinase |
| M. abscessus ATCC 19977 | MAB_3188c | - | 1e-114 | 88.89% (234) | uridylate kinase PyrH |
| M. marinum M | MMAR_1826 | pyrH | 1e-130 | 89.05% (274) | uridylate kinase PyrH |
| M. avium 104 | MAV_3733 | pyrH | 1e-120 | 96.51% (229) | uridylate kinase |
| M. smegmatis MC2 155 | MSMEG_2540 | pyrH | 1e-120 | 92.77% (235) | uridylate kinase |
| M. thermoresistible (build 8) | TH_0271 | pyrH | 1e-109 | 89.14% (221) | PROBABLE URIDYLATE KINASE PYRH (UK) (URIDINE MONOPHOSPHATE |
| M. ulcerans Agy99 | MUL_2074 | pyrH | 1e-131 | 89.05% (274) | uridylate kinase PyrH |
| M. vanbaalenii PYR-1 | Mvan_2220 | pyrH | 1e-119 | 92.34% (235) | uridylate kinase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_4124|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_2220|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_2540|M.smegmatis_MC2_155 --------------------------------------------------
TH_0271|M.thermoresistible__bu --------------------------------------------------
MAB_3188c|M.abscessus_ATCC_199 --------------------------------------------------
MLBr_01591|M.leprae_Br4923 --------------------------------------------------
MAV_3733|M.avium_104 --------------------------------------------------
Mb2907c|M.bovis_AF2122/97 --------------------------------------------------
Rv2883c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_1826|M.marinum_M --------------------------------------------------
MUL_2074|M.ulcerans_Agy99 MSGPGPPFQQACLVPSGAPRLGSTARARWRIPGAPAVQTSLRRCPRPDTG
Mflv_4124|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_2220|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_2540|M.smegmatis_MC2_155 --------------------------------------------------
TH_0271|M.thermoresistible__bu --------------------------------------------------
MAB_3188c|M.abscessus_ATCC_199 --------------------------------------------------
MLBr_01591|M.leprae_Br4923 -----------------------------------------MTESWESQV
MAV_3733|M.avium_104 --------------------------------------------------
Mb2907c|M.bovis_AF2122/97 -----------------------------------------MTEP---DV
Rv2883c|M.tuberculosis_H37Rv -----------------------------------------MTEP---DV
MMAR_1826|M.marinum_M -----------------------------------------MTES---DV
MUL_2074|M.ulcerans_Agy99 HAFGAVGYERTPRLPRSSAMRQDMRGRSGWLPGWHSHARRFMTES---DV
Mflv_4124|M.gilvum_PYR-GCK --MADP----VTDQTPESRR-----------------AYTRVLLKLGGEM
Mvan_2220|M.vanbaalenii_PYR-1 --MADPTAEPVAGQSSLIRP-----------------AYSRVLLKLGGEM
MSMEG_2540|M.smegmatis_MC2_155 --MADSN---VAGRAAPIRP-----------------LYTRVLLKLGGEM
TH_0271|M.thermoresistible__bu -------------------------------------------------M
MAB_3188c|M.abscessus_ATCC_199 ------------MTRDVSRT-----------------GYKRVLLKLGGEM
MLBr_01591|M.leprae_Br4923 AGSAVPKPQLMSANGVSAAAAGRGERLNHAGNPGHRSKYSRVLLKLGGEM
MAV_3733|M.avium_104 -----------------------------------------MLLKLGGEM
Mb2907c|M.bovis_AF2122/97 AGAPASKPEPASTGAASAAQ-----------LSG----YSRVLLKLGGEM
Rv2883c|M.tuberculosis_H37Rv AGAPASKPEPASTGAASAAQ-----------LSG----YSRVLLKLGGEM
MMAR_1826|M.marinum_M AGGAAPKPEPASANGVPAAP-----------VSGSVSQYKRVLLKLGGEM
MUL_2074|M.ulcerans_Agy99 AGGAAPKPEPASANGVPAAP-----------VSGSVSQYKRVLLKLGGEM
*
Mflv_4124|M.gilvum_PYR-GCK FGGGQVGLDPDVVALVARQIAEVVRSGAQVAVVIGGGNFFRGAQLQQRGM
Mvan_2220|M.vanbaalenii_PYR-1 FGGGQVGLDPDVVAQVARQIAEVVRSGAQVAVVIGGGNFFRGAQLQQRGM
MSMEG_2540|M.smegmatis_MC2_155 FGGGQVGLDPDVVAQVARQIAEVVRSGAQVAVVIGGGNFFRGAQLQQRGM
TH_0271|M.thermoresistible__bu FGGGQVGLDPDVVQKVAHEIAEVVRSGVQVAVVIGGGNFFRGAQLQQRGL
MAB_3188c|M.abscessus_ATCC_199 FGGGAVGLDPDVVHRVARQIAEVVRTGAEVAVVIGGGNFFRGAQLQQRGM
MLBr_01591|M.leprae_Br4923 FGGGQVGLDLDVVAQVARQIAEVVRGGVQVAVVIGGGNFFRGAQLQQRGM
MAV_3733|M.avium_104 FGGGQVGLDPDVVAQVARQIAEVVRGGVQVAVVIGGGNFFRGAQLQQRGM
Mb2907c|M.bovis_AF2122/97 FGGGQVGLDPDVVAQVARQIADVVRGGVQIAVVIGGGNFFRGAQLQQLGM
Rv2883c|M.tuberculosis_H37Rv FGGGQVGLDPDVVAQVARQIADVVRGGVQIAVVIGGGNFFRGAQLQQLGM
MMAR_1826|M.marinum_M FGGGQVGLDPDVVAQVARQIAEVVRGGVQVAVVIGGGNFFRGAQLQQRGM
MUL_2074|M.ulcerans_Agy99 FGGGQVGLDPDVVAQVARQIAEVVRGGVQVAVVIGGGNFFRGAQLQQRGM
**** **** *** **::**:*** *.::***************** *:
Mflv_4124|M.gilvum_PYR-GCK ERTRSDYMGMLGTVMNSLALQDFLQKEGIDTRVQTAITMGQVAEPYIPLR
Mvan_2220|M.vanbaalenii_PYR-1 DRARSDYMGMLGTVMNSLALQDFLQKEGIDTRVQTAITMGQVAEPYIPLR
MSMEG_2540|M.smegmatis_MC2_155 ERTRSDYMGMLGTVMNSLALQDFLQKEGIDTRVQTAITMGQVAEPYIPLR
TH_0271|M.thermoresistible__bu ERSRSDYMGMLGTVMNSLALQDFLQKEGIDTRVQTAITMGQVAEPYIPLR
MAB_3188c|M.abscessus_ATCC_199 ERTRSDYMGMLGTVMNSLALQDFLEKEGIVTRVQTAITMGQVAEPYIPLR
MLBr_01591|M.leprae_Br4923 ERTRSDYMGMLGTVMNSLALQDFLEKEGIATRVQTAITMGQVAEPYLPLR
MAV_3733|M.avium_104 ERTRSDYMGMLGTVMNSLALQDFLEKEGIVTRVQTAITMGQVAEPYLPLR
Mb2907c|M.bovis_AF2122/97 ERTRSDYMGMLGTVMNSLALQDFLEKEGIVTRVQTAITMGQVAEPYLPLR
Rv2883c|M.tuberculosis_H37Rv ERTRSDYMGMLGTVMNSLALQDFLEKEGIVTRVQTAITMGQVAEPYLPLR
MMAR_1826|M.marinum_M ERTRSDYMGMLGTVMNSLALQDFLEKEGIATRVQTAITMGQVAEPYLPLR
MUL_2074|M.ulcerans_Agy99 ERTRSDYMGMLGTVMNSLALQDFLEKEGIATRVQTAITMGQVAEPYLPLR
:*:*********************:**** ****************:***
Mflv_4124|M.gilvum_PYR-GCK AVRHLEKGRVVIFGAGMGLPYFSTDTTAAQRALEIGAEVVLMAKAVDGIY
Mvan_2220|M.vanbaalenii_PYR-1 AVRHLEKGRVVIFGAGMGLPYFSTDTTAAQRALEIGADVVLMAKAVDGVF
MSMEG_2540|M.smegmatis_MC2_155 AVRHLEKGRVVIFGAGMGLPYFSTDTTAAQRALEIGAEVVLMAKAVDGVF
TH_0271|M.thermoresistible__bu AQRHLEKGRVVIFGAGMGLPYFSTDTTAAQRALEIGAEVVLMAKAVDGVY
MAB_3188c|M.abscessus_ATCC_199 ARRHLEKGRVVIFGAGMGLPYFSTDTTAAQRALEIGAEVVLMAKAVDGIF
MLBr_01591|M.leprae_Br4923 AVRHLEKGRVVIFGAGMGLPYFSTDTTAAQRALEIGADVVLMAKAVDGVF
MAV_3733|M.avium_104 AVRHLEKGRVVIFGAGMGLPYFSTDTTAAQRALEIGAEVVLMAKAVDGVF
Mb2907c|M.bovis_AF2122/97 AVRHLEKGRVVIFGAGMGLPYFSTDTTAAQRALEIGADVVLMAKAVDGVF
Rv2883c|M.tuberculosis_H37Rv AVRHLEKGRVVIFGAGMGLPYFSTDTTAAQRALEIGADVVLMAKAVDGVF
MMAR_1826|M.marinum_M AVRHLEKGRVVIFGAGMGLPYFSTDTTAAQRALEIGADVVLMAKAVDGVF
MUL_2074|M.ulcerans_Agy99 AVRHLEKGRVVIFGAGMGLPYFSTDTTAAQRALEIGADVVLMAKAVDGVF
* ***********************************:**********::
Mflv_4124|M.gilvum_PYR-GCK TDDPRLNPAAEMLTAISHREVIDRGLKVADATAFSLCMDNGMPILVFNLL
Mvan_2220|M.vanbaalenii_PYR-1 TADPRVDPDAELLIAISHREVIDRGLKVADATAFSLCMDNGMPILVFNLL
MSMEG_2540|M.smegmatis_MC2_155 TDDPRTNPDAELITAISHREVIDRGLKVADATAFSLCMDNGMPILVFNLL
TH_0271|M.thermoresistible__bu TDDPRTNPDAKLITKITHREVIDRGLRVADATAFSLCMDNGMPILVFNLL
MAB_3188c|M.abscessus_ATCC_199 TDDPRANPDAEFLPEITHREVIDRGLQVADATAFSLCMDNRMPILVFNLL
MLBr_01591|M.leprae_Br4923 VEDPRVNPEAELLTAISHREVIDRGLRVADATAFSLCMDNGMPILVFNLL
MAV_3733|M.avium_104 SADPRQYPEAELITAISHREVIDRGLRVADATAFSLCMDNGMPILVFNLL
Mb2907c|M.bovis_AF2122/97 AEDPRVNPEAELLTAVSHREVLDRGLRVADATAFSLCMDNGMPILVFNLL
Rv2883c|M.tuberculosis_H37Rv AEDPRVNPEAELLTAVSHREVLDRGLRVADATAFSLCMDNGMPILVFNLL
MMAR_1826|M.marinum_M AEDPRVNPHAELLAVISHREVIDRGLRVADATAFSLCMDNGMPILVFNLL
MUL_2074|M.ulcerans_Agy99 AEDPRVNPHAELLAVISHREVIDRGLRVADATAFSLCMDNGMPILVFNLL
*** * *::: ::****:****:************* *********
Mflv_4124|M.gilvum_PYR-GCK TDGNIARAVAGERIGTLVTT
Mvan_2220|M.vanbaalenii_PYR-1 VDGNIARAVAGEKIGTLVTT
MSMEG_2540|M.smegmatis_MC2_155 TSGNIARAVAGEKIGTLVTT
TH_0271|M.thermoresistible__bu IDGNIARAVAGEKIGTLVTT
MAB_3188c|M.abscessus_ATCC_199 TEGNIARAVAGEKIGTLVTP
MLBr_01591|M.leprae_Br4923 TNGNIARAVAGEKIGTLVTT
MAV_3733|M.avium_104 TNGNIARAVAGEKIGTLVTT
Mb2907c|M.bovis_AF2122/97 TDGNIARAVRGEKIGTLVTT
Rv2883c|M.tuberculosis_H37Rv TDGNIARAVRGEKIGTLVTT
MMAR_1826|M.marinum_M TNGNIARAVAGEKIGTLVTT
MUL_2074|M.ulcerans_Agy99 TNGNIARAVAGEKIGTLVTT
.******* **:******.