For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VLAENILMAGGVDSIGRISVLDLVPVRTDQSSRHALVATVQLTQTANQRNFSSRLVVEDYSMLSVGATSP PVLITYLAVQTTRIELLGNLGAVSERASFNYTRSVSRARIDLG
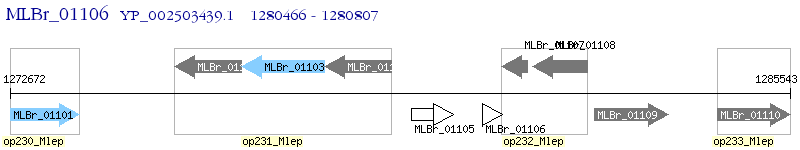
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. leprae Br4923 | MLBr_01106 | - | - | 100% (113) | hypothetical protein MLBr_01106 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_2237 | - | 4e-15 | 43.69% (103) | luciferase family protein |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_3639 | - | 5e-06 | 27.45% (102) | putative luciferase-like protein |
| M. marinum M | MMAR_4179 | - | 5e-21 | 52.43% (103) | monooxygenase |
| M. avium 104 | MAV_1409 | - | 3e-19 | 49.51% (103) | alkanal monooxygenase alpha chain |
| M. smegmatis MC2 155 | MSMEG_5029 | - | 7e-16 | 43.69% (103) | alkanal monooxygenase alpha chain |
| M. thermoresistible (build 8) | TH_4625 | - | 4e-18 | 44.95% (109) | PUTATIVE alkanal monooxygenase alpha chain |
| M. ulcerans Agy99 | MUL_4481 | - | 4e-21 | 52.43% (103) | monooxygenase |
| M. vanbaalenii PYR-1 | Mvan_4458 | - | 7e-15 | 41.75% (103) | luciferase family protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_2237|M.gilvum_PYR-GCK ---------------MRLSVLDLVPVRSDQSTGDALAATTLLAQAADRLG
Mvan_4458|M.vanbaalenii_PYR-1 --------------MMRLSVLDLVPVRSDQSTSDAIAASTLLAQTADRLG
MSMEG_5029|M.smegmatis_MC2_155 ---------------MRLSVLDLVPVRTDQTTADALAATTHLAQTADRLG
TH_4625|M.thermoresistible__bu -----VTVMPGVDPDMRLSVLDLVPVRTDQTTADALTASTQLAQTADRLG
MMAR_4179|M.marinum_M ---------------MRLSVLDLVPVRTDQSTRDALAATVSLAQTAERLG
MUL_4481|M.ulcerans_Agy99 ------------MGSVRLSVLDLVPVRTDQSTRDALAATVSLAQTAERLG
MAV_1409|M.avium_104 ---------------MRLSVLDLVPVRTDQTTGDALAATVALAQAADRLG
MLBr_01106|M.leprae_Br4923 MLAENILMAGGVDSIGRISVLDLVPVRTDQSSRHALVATVQLTQTANQRN
MAB_3639|M.abscessus_ATCC_1997 -------------MARPVSILDLARVAPHETVAESFAASVEIARHAETLG
:*:***. * ..:: .::.*:. ::: *: .
Mflv_2237|M.gilvum_PYR-GCK YTRYWVAEHHNMPAVAATSPPVLIAHLAAHTAAIRLGSGGVMLPNHAPLA
Mvan_4458|M.vanbaalenii_PYR-1 YTRYWVAEHHNMPSVAATSPPVLIAHLAAHTASMRVGSGGVMLPNHAPLA
MSMEG_5029|M.smegmatis_MC2_155 YHRFWVAEHHNMPAVAATSPPVLIAHLAAHTSQLRLGSGGVMLPNHAPLA
TH_4625|M.thermoresistible__bu YTRYWVAEHHNMEAVAATSPPVLIAHLAAHTTRMRIGSGGVMLPNHAPLA
MMAR_4179|M.marinum_M FTRYWVAEHHNMASVGATSPPVLIAYLAAQTTHLRLGSGGVMLPNHAPLA
MUL_4481|M.ulcerans_Agy99 FTRYWVAEHHNMASVGATSPPVLIAYLAAQTTHLRLGSGGVMLPNHAPLA
MAV_1409|M.avium_104 FTRYWVAEHHNMPSVGATSPPVLLAYLAPQTTRLRLGSGGVMLPNHAPLA
MLBr_01106|M.leprae_Br4923 FSSRLVVEDYSMLSVGATSPPVLITYLAVQTTRIEL------LGNLGAVS
MAB_3639|M.abscessus_ATCC_1997 FHRVWYAEHHNMRSIASSATSVLIAHVATQTSTIRLGAGGIMLPNHSPLQ
: .*.:.* ::.:::..**::::* :*: :.: * * ..:
Mflv_2237|M.gilvum_PYR-GCK VAEQFALLEAAHPGRIDLGIGRAPGSDPVTSLALRGAAG--RDDRDIEAF
Mvan_4458|M.vanbaalenii_PYR-1 VAEQFALLEAAHPGRIDLGIGRAPGSDPVTSLALRGAAG--RDERDIEAF
MSMEG_5029|M.smegmatis_MC2_155 VAEQFALLEAAHPGRIDLGIGRAPGSDPVTSLALRGAAG--RDDRDIEAF
TH_4625|M.thermoresistible__bu VAEQFALLEAAHPGRIDLGIGRAPGSDPVTSLALRGAAG--RDDRDIEAF
MMAR_4179|M.marinum_M VAEQFALLEAAAPGRIDLGIGRAPGSDPVTSYALRGGRSPAQQERDIESF
MUL_4481|M.ulcerans_Agy99 VAEQFALLEAAAPGRIDLGIGRAPGSDPVTSYALRGGRSPAQQERDIESF
MAV_1409|M.avium_104 VAEQFALLEAAAPGRIDLGIGRAPGSDPVTSYALRGGRG----DEDIQRF
MLBr_01106|M.leprae_Br4923 ERASFNYTRSVSRARIDLG-------------------------------
MAB_3639|M.abscessus_ATCC_1997 IAEQFGTLETLHPGRIDLGLGRAPGTDQVTLRALRHGPG------DAEHF
.* .: .*****
Mflv_2237|M.gilvum_PYR-GCK PEYLDDVMALMSSKGVRVPLPRDLMRDNYILKATPAAASTPRLWLLGSSM
Mvan_4458|M.vanbaalenii_PYR-1 PQYLDDVVALMSSKGVRVPLPRDLMRDNYILKATPAAVTEPRLWLLGSSM
MSMEG_5029|M.smegmatis_MC2_155 PQYLDDVVALMGARGVRVPLPRDLLRDNYILKATPNATSVPQLWLLGSSM
TH_4625|M.thermoresistible__bu PDHLDDIVALMSPHGVRVPIP----QQRYILKATPKAVSVPQLWLLGSST
MMAR_4179|M.marinum_M PDYLDDVVALMSARGVRVPLQ----SGDYILKATPAALTQPRLWLLGSSM
MUL_4481|M.ulcerans_Agy99 PDYLDDVVALMSARGARVPLQ----SGDYILKATSAALTQPRLWLLGSSM
MAV_1409|M.avium_104 PDYLDDVAALMSARGVVVPLR----SGDYRLRATPAAAGEPRLWLLGSSM
MLBr_01106|M.leprae_Br4923 --------------------------------------------------
MAB_3639|M.abscessus_ATCC_1997 P---QDVLELQAFLGDQSRVPG--------VTATPGHGTQVPLYILGSSL
Mflv_2237|M.gilvum_PYR-GCK YSARLAAAKGLPYVFAHHFAGQGTAEALALYRENFQPSEQTPEPVTFLTV
Mvan_4458|M.vanbaalenii_PYR-1 YSAQLAAAKGLPYVFAHHFAGQGTEEALQYYRDNFRPSDLTPEPVTFLTV
MSMEG_5029|M.smegmatis_MC2_155 YSAHLAAAKGLPYVFAHHFSGHGTAEALAIYREEFQPSEAAPEPVTFLTV
TH_4625|M.thermoresistible__bu YSAHLAAAKGLPYVFAHHFSGQGTAEALRIYRENFRPGKHTPEPLTFLTV
MMAR_4179|M.marinum_M YSARLAAAKGLPYVFAHHFSGKGTEEALELYRSRFVPSALAARPVTFLTV
MUL_4481|M.ulcerans_Agy99 YSARLAAAKGLPYVFAHHFSGKGTEEALELYRSRFVPSALAARPVTFLTV
MAV_1409|M.avium_104 YSAHLAAAKGLPYVFAHHFSGKGTEEALEVYRSRFRPSALTPEPLTFLTV
MLBr_01106|M.leprae_Br4923 --------------------------------------------------
MAB_3639|M.abscessus_ATCC_1997 FGAKLAAVLGLPYGFASHFAPPALQDAVALYRREFQPSQQLDAPYVLAGV
Mflv_2237|M.gilvum_PYR-GCK NAVVAESHDEAMRLVLPNLHMMARLRTGQPLVPLDLVEDAEALDLGPRAQ
Mvan_4458|M.vanbaalenii_PYR-1 NAAVAETHDDAMRLILPNLQMMARLRTGQPMVPIDLVEDAEAQVVSPRAQ
MSMEG_5029|M.smegmatis_MC2_155 NAVVAETTEEAEALLLPQLQMMARLRTGQPLNALDLVEDAQTTVLTPQGD
TH_4625|M.thermoresistible__bu NAVVADTRDEAMALLLPQLQMMARLRTGEPLGALDLVEDAEQRPLTPQQR
MMAR_4179|M.marinum_M NAAVADTREEATALMMPNLHMMARLRTGQPLGAVQLVEEAQDARLTPEQQ
MUL_4481|M.ulcerans_Agy99 NAAVADTREEATALMMPNLHMMARLRTGQPLGPVQLVEEAQDARLTPEQQ
MAV_1409|M.avium_104 NAVVAETRDEATALMLPNLQMMAKLRTGQPLGPVPLVEDAEHAELTAEQR
MLBr_01106|M.leprae_Br4923 --------------------------------------------------
MAB_3639|M.abscessus_ATCC_1997 NVIAADDEATARDQHQAALIERAKLFLHRSGGPR-LTDEEALAALESPAG
Mflv_2237|M.gilvum_PYR-GCK AVIDGGRNLAVAGAPAEAADQLLSLAEEFGVDEVMVNPVASAYRGVDPAT
Mvan_4458|M.vanbaalenii_PYR-1 AVIDAGLRQAVVGSPAEAGDQLRALAEKFGVDEVMVHPVASAFRGVDPAT
MSMEG_5029|M.smegmatis_MC2_155 AVVADGRRRAVVGSPTEAAEQVRALAEEFDVDEVMINPVASAHRGADPRT
TH_4625|M.thermoresistible__bu AVVEAGARXXXX--RTRRAGSVRPRPR-----------------------
MMAR_4179|M.marinum_M RIVDSGLERAVVGSPEEAAEQVRALGQRFGVDEVMVHPVASAHRGTDPAT
MUL_4481|M.ulcerans_Agy99 RIVDSGLERAVVGSPEEAAEQVRALGQRFGVDEVMVHPVASAHRGTGPAT
MAV_1409|M.avium_104 RIVDSGLRRAVLGGPAEAAEQVRALAERFGVDEVMVNPVASARRGTDPAT
MLBr_01106|M.leprae_Br4923 --------------------------------------------------
MAB_3639|M.abscessus_ATCC_1997 RQIQSMTKYFAVGTADQVGAYLEDFAQLADADELIVVPNAPDRKLQLRSL
Mflv_2237|M.gilvum_PYR-GCK APAREATLELLAKELF
Mvan_4458|M.vanbaalenii_PYR-1 APAREATLELLAKEIF
MSMEG_5029|M.smegmatis_MC2_155 ATARDTTLELLAKELF
TH_4625|M.thermoresistible__bu ----------------
MMAR_4179|M.marinum_M APARVATCELLAKELF
MUL_4481|M.ulcerans_Agy99 APARVATCELLAKELF
MAV_1409|M.avium_104 APARVATLELLAKELF
MLBr_01106|M.leprae_Br4923 ----------------
MAB_3639|M.abscessus_ATCC_1997 EIVAEVSGLTLRG---