For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VNRRILTLLVALVPILVFGVLLAVVTVPFVSLGPGPTFDTLGEVDGKQVVAIKGAHTHPTSGHLNMTTVS QRDDLTLGEALTLWLSGQEQLVPRDLIYPPGKSREDVDKVNYADFKQSEDSAAYAALGYLQYPSAVTVAK VTDPGQSVGKLKSGDAVDAVNGSPVANVEQFTGLLKKTKPGEVVTIDFRRKNEPPGVVQITLGANKDRDY GFLGVAVLDAPWAPFVVDFNLANVGGPSAGLMFSLAVVDKLTTGDLVGSTFVAGTGTISIDGKVGPIGGI AHKMAAARAAGATVFLVPAKNCYEARSDNRTGLRLVKVESLSQAVDALHAMTSGGQLPSC
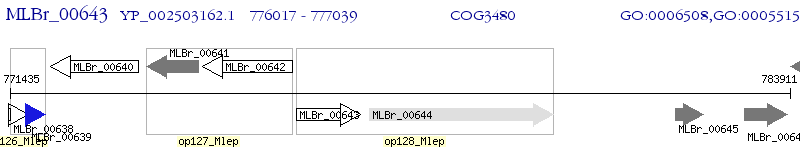
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. leprae Br4923 | MLBr_00643 | - | - | 100% (340) | putative secreted protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3216c | - | 1e-159 | 80.59% (340) | hypothetical protein Mb3216c |
| M. gilvum PYR-GCK | Mflv_4655 | - | 1e-135 | 67.35% (340) | PDZ/DHR/GLGF |
| M. tuberculosis H37Rv | Rv3194c | - | 1e-159 | 80.59% (340) | hypothetical protein Rv3194c |
| M. abscessus ATCC 19977 | MAB_3499c | - | 1e-134 | 67.35% (340) | hypothetical protein MAB_3499c |
| M. marinum M | MMAR_1370 | - | 1e-165 | 84.12% (340) | serine protease |
| M. avium 104 | MAV_4138 | - | 1e-170 | 87.35% (340) | PDZ domain-containing protein |
| M. smegmatis MC2 155 | MSMEG_1958 | - | 1e-143 | 72.51% (342) | PDZ domain-containing protein |
| M. thermoresistible (build 8) | TH_1131 | - | 1e-138 | 68.53% (340) | POSSIBLE CONSERVED SECRETED PROTEIN |
| M. ulcerans Agy99 | MUL_2506 | - | 1e-166 | 84.71% (340) | serine protease |
| M. vanbaalenii PYR-1 | Mvan_1813 | - | 1e-135 | 68.24% (340) | PDZ/DHR/GLGF |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_4655|M.gilvum_PYR-GCK -------------------MNRRISTLVVALAPILIFGILLSAVTVPYVA
Mvan_1813|M.vanbaalenii_PYR-1 MNSGGLRGSPRALTSTVGRVNRRISTMVVALVPILVFGVLLTVVTVPYVA
MSMEG_1958|M.smegmatis_MC2_155 ----------------MNGVNRRILTLLVALVPVVAFGVLLSVVQVPFVS
TH_1131|M.thermoresistible__bu -------------------VNRRILTLIVALVPIMAFGVLLSVVRVPFVS
Mb3216c|M.bovis_AF2122/97 -------------------MNRRILTLMVALVPIVVFGVLLAVVTVPFVA
Rv3194c|M.tuberculosis_H37Rv -------------------MNRRILTLMVALVPIVVFGVLLAVVTVPFVA
MLBr_00643|M.leprae_Br4923 -------------------MNRRILTLLVALVPILVFGVLLAVVTVPFVS
MAV_4138|M.avium_104 -------------------MNRRILTLMVALVPIVAFGVLLAVVTVPFVS
MMAR_1370|M.marinum_M -------------------MNRRILTLMVALVPIVVFGVLLAVVTVPFVS
MUL_2506|M.ulcerans_Agy99 -------------------MNRRILTLMVALVPIVVFGVLLAVVTVPFVS
MAB_3499c|M.abscessus_ATCC_199 -------------------MNRRVATLLVALIPIVVFGLLMSVITVPFVS
:***: *::*** *:: **:*::.: **:*:
Mflv_4655|M.gilvum_PYR-GCK LGPGPTFDTLGEVDGKEVVDIEG--ADVHATSGHLNMTTVSQRDGLTLGQ
Mvan_1813|M.vanbaalenii_PYR-1 LGPGPTFDTLGEVDGKQVVDIEG--AHVHKTSGHLNMTTVSQRDGLTLGQ
MSMEG_1958|M.smegmatis_MC2_155 LGPGPTFNTLGEIDGKQVVDIEGPDATVHPTSGHLNMTTVSQRDGLTLGQ
TH_1131|M.thermoresistible__bu LGPGPTFDTLGQIDGKDVVAIEG--AEVYPTSGQLNMTTVAQRDGLTLGQ
Mb3216c|M.bovis_AF2122/97 LGPGPTFDTLGEIDGKQVVQIVG--TQTYPTSGHLNMTTVSQRDGLTLGE
Rv3194c|M.tuberculosis_H37Rv LGPGPTFDTLGEIDGKQVVQIVG--TQTYPTSGHLNMTTVSQRDGLTLGE
MLBr_00643|M.leprae_Br4923 LGPGPTFDTLGEVDGKQVVAIKG--AHTHPTSGHLNMTTVSQRDDLTLGE
MAV_4138|M.avium_104 LGPGPTFDTLGEVDGKQVVAIEG--TMTHPTTGHLNMTTVSQRDDLTLGE
MMAR_1370|M.marinum_M LGPGPTFDTLGEVDGRQVVQIEG--TQTHPTTGHLNMTTVSQRDGLSLGE
MUL_2506|M.ulcerans_Agy99 LGPGPTFDTLGEVDGKQVVQIEG--TQTHPTTGHLNMTTVSQRDGLSLGE
MAB_3499c|M.abscessus_ATCC_199 LGPGPTFNTLGQVEGKEVVDIKG--TKVDPVSGHLNMTTVSQRDGLTLAD
*******:***:::*::** * * : . .:*:******:***.*:*.:
Mflv_4655|M.gilvum_PYR-GCK ALIFWASGRDQLLPRDLVYPPDKTREEIDEANSQDFRQSEDSAEYAALHH
Mvan_1813|M.vanbaalenii_PYR-1 ALVFWASGRDQLVPRELVYPPDKSREEIDEANTRDFRDSEDSAEYAALQY
MSMEG_1958|M.smegmatis_MC2_155 AITLWMSGREQLVPRDLVYPPDKSKDEIDEANTADFKNSEDSAEYAALSF
TH_1131|M.thermoresistible__bu ALTLWMSGREQLVPRDLVYPPDKSREEIEQANTRDFRQSEDSAEYAALSY
Mb3216c|M.bovis_AF2122/97 ALALWLSGQEQLMPRDLVYPPGKSREEIENDNAADFKRSEAAAEYAALGY
Rv3194c|M.tuberculosis_H37Rv ALALWLSGQEQLMPRDLVYPPGKSREEIENDNAADFKRSEAAAEYAALGY
MLBr_00643|M.leprae_Br4923 ALTLWLSGQEQLVPRDLIYPPGKSREDVDKVNYADFKQSEDSAAYAALGY
MAV_4138|M.avium_104 ALTLWLSDTEQLVPRDLIYPPGKSREDVDKANNADFKQSEDSAAYAALGY
MMAR_1370|M.marinum_M ALTLWLSGQEQLVPRDLVYPPGKSREEVDNANSADFKNSEESAEYAALEY
MUL_2506|M.ulcerans_Agy99 ALTLWLSGQEQLVPRDLVYPPGKSREEVDNANSADFKNSEESAEYAALEY
MAB_3499c|M.abscessus_ATCC_199 ALRLWASGSEQLVPRDVVYPPDRSKDQVDKENDLEFKKSEESAEFAALGY
*: :* *. :**:**:::***.:::::::: * :*: ** :* :*** .
Mflv_4655|M.gilvum_PYR-GCK LKYPMAVTVESIDEEGPSKGKLQGGDAIDGVNGKPVATLDQFQELLKAYK
Mvan_1813|M.vanbaalenii_PYR-1 LKYPMAVTVESIDEKGPSKGKLQAGDAIDAVNNTPVASLAQFQELLKSTK
MSMEG_1958|M.smegmatis_MC2_155 LKYPMAVTVQNVTDPGPSAGKLQDGDAIDGVNGKPVANLDEFQALLKETK
TH_1131|M.thermoresistible__bu LDYPMAVTVDSVEESGPSAGKLRDGDAIDAVDGRPVANLDEFQAVLQETK
Mb3216c|M.bovis_AF2122/97 LKYPKAVTVASVMDPGPSVDKLQAGDAIDAVDGTPVGNLDQFTALLKNTK
Rv3194c|M.tuberculosis_H37Rv LKYPKAVTVASVMDPGPSVDKLQAGDAIDAVDGTPVGNLDQFTALLKNTK
MLBr_00643|M.leprae_Br4923 LQYPSAVTVAKVTDPGQSVGKLKSGDAVDAVNGSPVANVEQFTGLLKKTK
MAV_4138|M.avium_104 LKYPSAVTVAKVPEPGPSAGKLKVGDAIDAVNGTPVATVEEFTGLLKNTK
MMAR_1370|M.marinum_M LKYPDAVTVATVSEPGPSAGKLKAGDAIDAVNGTAVANVEQFTSILKDTK
MUL_2506|M.ulcerans_Agy99 LKYPDAVTVATVSEPGPSAGKLKAGDAIDAVNGTAVANVEQFTSILKDTK
MAB_3499c|M.abscessus_ATCC_199 LKYPAAVTVLTISDDGPSKGKLQQNDAITAVNGHPVATLDEFTGQLKLTK
*.** **** .: : * * .**: .**: .*:. .*..: :* *: *
Mflv_4655|M.gilvum_PYR-GCK PGDTIAVDYRRKNAPMGTAEITLGKHPDRDQGILGVNVLDAPWAPFTIDF
Mvan_1813|M.vanbaalenii_PYR-1 PGDTIIVDYRRKNAPMGTTEITLGSHPDRPQGVVGVNVLDAPWAPFTIDF
MSMEG_1958|M.smegmatis_MC2_155 PGDKLVIDYRRKNAPPGEATITLGDNPDRDYGYLGIAVLDAPWAPFDIEF
TH_1131|M.thermoresistible__bu PGDTIVIDYRRKNAPPGAVAVTLGENPDREHGYLGIGVLDAPWAPFEITF
Mb3216c|M.bovis_AF2122/97 PGQEVTIDFRRKNEPPGIAQITLGKNKDRDQGVLGIEVVDAPWAPFAVDF
Rv3194c|M.tuberculosis_H37Rv PGQEVTIDFRRKNEPPGIAQITLGKNKDRDQGVLGIEVVDAPWAPFAVDF
MLBr_00643|M.leprae_Br4923 PGEVVTIDFRRKNEPPGVVQITLGANKDRDYGFLGVAVLDAPWAPFVVDF
MAV_4138|M.avium_104 PGQTVTIDYRRKNEPAGVAQITLGANKDRDYGFLGVAVLDAPWAPFVVNF
MMAR_1370|M.marinum_M PGDVVVIDFRRKNEAPGVAQITLGTNKDRDYGFMGVAVLDAPWAPFVVDF
MUL_2506|M.ulcerans_Agy99 PGDVVVIDFRRKNEAPGVAQITLGTNKDRDYGFMGVAVLDAPWAPFVVDF
MAB_3499c|M.abscessus_ATCC_199 PGDEVMLDYKRKNAPNGTVKITLGKNGDRDYGFLGVGVVQAPWAPFTIDF
**: : :*::*** . * . :*** : ** * :*: *::****** : *
Mflv_4655|M.gilvum_PYR-GCK NLANIGGPSAGLMFSLAVVDKLTTGDVTGGKFVAGSGTISGDGKVGSIGG
Mvan_1813|M.vanbaalenii_PYR-1 NLANIGGPSAGLMFSLAVVDKLTTGDVNGGKFVAGTGTISGDGKVGSIGG
MSMEG_1958|M.smegmatis_MC2_155 NLANIGGPSAGLMFSLAVVDKLTTGDLNDGKFIAGTGTISGDGKVGSIGG
TH_1131|M.thermoresistible__bu NLANVGGPSAGMVFSLAVIDKLTEGDLAGSRVVAGSGTITGDGQVGSIGG
Mb3216c|M.bovis_AF2122/97 HLANVGGPSAGLMFSLAVVDKLTSGHLVGSTFVAGTGTIAVDGKVGQIGG
Rv3194c|M.tuberculosis_H37Rv HLANVGGPSAGLMFSLAVVDKLTSGHLVGSTFVAGTGTIAVDGKVGQIGG
MLBr_00643|M.leprae_Br4923 NLANVGGPSAGLMFSLAVVDKLTTGDLVGSTFVAGTGTISIDGKVGPIGG
MAV_4138|M.avium_104 NLANVGGPSAGLMFSLAVVDKLTTGDLAGSNFVAGTGTISADGKVGQIGG
MMAR_1370|M.marinum_M HLANVGGPSAGLMFSLAVVDKLTTGDLVGSTFVAGTGTITADGKVGPIGG
MUL_2506|M.ulcerans_Agy99 HLANVGGPSAGLMFSLAVVDKLTTGDLVGSTFVAGTGTITADGKVGPIGG
MAB_3499c|M.abscessus_ATCC_199 NLANIGGPSAGLMFSLAVIDKLTPGELDGGKFVAGTGTIDAEGKVGPIGG
:***:******::*****:**** *.: .. .:**:*** :*:** ***
Mflv_4655|M.gilvum_PYR-GCK ITHKVTAAQEAGATVFLVPADNCAEARTADADGIELLKVETLEQAVDGLR
Mvan_1813|M.vanbaalenii_PYR-1 ITHKIVAARDAGATAFLVPADNCAEAKTADADGIELLKVDTLEHAVDGLR
MSMEG_1958|M.smegmatis_MC2_155 ITHKMLAASEAGAEVFLVPADNCTEARSAPRDGLELVKVETLEGAVDALK
TH_1131|M.thermoresistible__bu ITHKMAAAADAGATVFLVPAENCVEAKSAQRDGMELVRVETLDDAVTALN
Mb3216c|M.bovis_AF2122/97 ITHKMAAARAAGATVFLVPAKNCYEASSDSPPGLKLVKVETLSQAVDALH
Rv3194c|M.tuberculosis_H37Rv ITHKMAAARAAGATVFLVPAKNCYEASSDSPPGLKLVKVETLSQAVDALH
MLBr_00643|M.leprae_Br4923 IAHKMAAARAAGATVFLVPAKNCYEARSDNRTGLRLVKVESLSQAVDALH
MAV_4138|M.avium_104 ITHKMVAAHAAGATVFLVPAKNCYEASSDNPSGLRLVKVETLAQAVDALH
MMAR_1370|M.marinum_M ITHKMAAARAAGASVFLVPAKNCYEAASDTPSGLRLVKVETLGQAVDALH
MUL_2506|M.ulcerans_Agy99 ITHKMAAARAAGASVFLVPAKNCYEAASDTPSGLRLVKVETLGQAVDALH
MAB_3499c|M.abscessus_ATCC_199 ITHKMFAAKDAGATVFLVPAANCAEAKTDGGQGLTLVKVGTLTEAVDGLN
*:**: ** *** .***** ** ** : *: *::* :* ** .*.
Mflv_4655|M.gilvum_PYR-GCK TLSAGGEPPRC
Mvan_1813|M.vanbaalenii_PYR-1 TLSAGGEPPRC
MSMEG_1958|M.smegmatis_MC2_155 TISAGGEPPRC
TH_1131|M.thermoresistible__bu TLSAGGEPPRC
Mb3216c|M.bovis_AF2122/97 AMTSGSPTPSC
Rv3194c|M.tuberculosis_H37Rv AMTSGSPTPSC
MLBr_00643|M.leprae_Br4923 AMTSGGQLPSC
MAV_4138|M.avium_104 AITAGGQPPSC
MMAR_1370|M.marinum_M AMTAGTATPSC
MUL_2506|M.ulcerans_Agy99 AMTSGTATPSC
MAB_3499c|M.abscessus_ATCC_199 ALNRGEQPPSC
::. * * *