For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MRPRPAPAAAIALLATAVYAVMWIGYRHGWGWLARMDWSLLNGTRDLAVKHPGWLRAAEVVSVVLGPGTL AVLGIAVTLCAVVLRRLRAALVLALACAPCNELVTAGAKALVDRPRPPTALVGAASTSFPSGHALEATAG LLAMTLFALPMLNRVLRGLLVAAVAVALPAVGLSRVMLNVHYPSDVLAGWSLGYLYFLLCLLVFRPSAPD RSG*
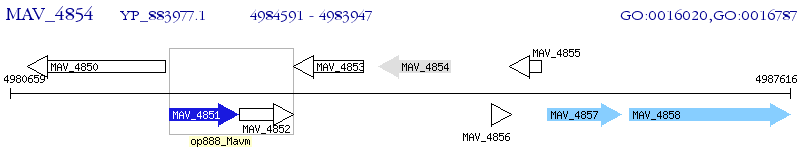
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_4854 | - | - | 100% (214) | PAP2 superfamily protein |
| M. avium 104 | MAV_3216 | - | 9e-17 | 37.35% (166) | PAP2 superfamily protein |
| M. avium 104 | MAV_0210 | - | 3e-08 | 35.48% (124) | PAP2 superfamily protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0316 | - | 9e-61 | 53.37% (208) | integral membrane protein |
| M. gilvum PYR-GCK | Mflv_0312 | - | 2e-39 | 42.13% (197) | phosphoesterase, PA-phosphatase related |
| M. tuberculosis H37Rv | Rv0308 | - | 9e-61 | 53.37% (208) | integral membrane protein |
| M. leprae Br4923 | MLBr_00094 | - | 4e-07 | 33.77% (154) | hypothetical protein MLBr_00094 |
| M. abscessus ATCC 19977 | MAB_0200c | - | 2e-38 | 41.58% (202) | phosphoesterase, PA-phosphatase related protein |
| M. marinum M | MMAR_0558 | - | 1e-59 | 55.77% (208) | hypothetical protein MMAR_0558 |
| M. smegmatis MC2 155 | MSMEG_0634 | - | 3e-44 | 47.26% (201) | PAP2 superfamily protein |
| M. thermoresistible (build 8) | TH_3242 | - | 1e-38 | 43.78% (201) | PUTATIVE putative phosphoesterase, PA-phosphatase related |
| M. ulcerans Agy99 | MUL_1222 | - | 8e-60 | 56.25% (208) | integral membrane protein |
| M. vanbaalenii PYR-1 | Mvan_0429 | - | 1e-45 | 45.96% (198) | phosphoesterase, PA-phosphatase related |
CLUSTAL 2.0.9 multiple sequence alignment
Mb0316|M.bovis_AF2122/97 ---MTRPQALLAVSLAFVATAVYAVMWVGHSQDWGWLHSFDWSLLNAAHD
Rv0308|M.tuberculosis_H37Rv ---MTRPQALLAVSLAFVATAVYAVMWVGHSQDWGWLHSFDWSLLNAAHD
MMAR_0558|M.marinum_M ---MTRPRMTLPLLLALAAIVVYALMWIGYRQDWGWLHAMDWSLLNASHD
MUL_1222|M.ulcerans_Agy99 ---MTRPRMTLPLLLALAAIVVYALMWIGYRQDWGWLHAMDWSLLNASHD
MAV_4854|M.avium_104 ---MMRPRPAPAAAIALLATAVYAVMWIGYRHGWGWLARMDWSLLNGTRD
MSMEG_0634|M.smegmatis_MC2_155 ----MERTRGLLIASALVAVAVYALMWIGYRSGWPWLVSFDDAGLDPAFR
Mvan_0429|M.vanbaalenii_PYR-1 ----MR--SGWLIGSAAAALGVYLLMWVGYVQGWVWLAGIDADVLNPANR
Mflv_0312|M.gilvum_PYR-GCK MTPDNRR----LLAGVVAALAVYGLLWAGWAQDWGWLTRADDAALDAAHR
MAB_0200c|M.abscessus_ATCC_199 MPARSDRGYVWPLLSAAAALIVFALLWLGYAQQWNGLSRLDVGALTPAHH
TH_3242|M.thermoresistible__bu ----VNRRTVGLIGSAVAAVVVYGLLWFATAHGWDRLARLDAAALSAAHD
MLBr_00094|M.leprae_Br4923 ---MPEDKAPTGELAAIAAVQSVLVDRPGVLPTARGMSHFGEHSIG-WLA
. * : . : . :
Mb0316|M.bovis_AF2122/97 IGIKNPAWVRFWDGVSLILGPVVLRPLGLLAAMVALAKRKIRIALLL-LA
Rv0308|M.tuberculosis_H37Rv IGIKNPAWVRFWDGVSLILGPVVLRPLGLLAAMVALAKRKIRIALLL-LA
MMAR_0558|M.marinum_M IGVKHPGWVNFWFAVSFVLGPITTRIVGLVAITIALVQRKMRMALLL-LA
MUL_1222|M.ulcerans_Agy99 IGVKHPGWVNFWFAVSFVLGPITTRIVGLVAITIALVQRKMRMALLL-LA
MAV_4854|M.avium_104 LAVKHPGWLRAAEVVSVVLGPGTLAVLGIAVTLCAVVLRRLRAALVLALA
MSMEG_0634|M.smegmatis_MC2_155 FGTEHPAWVTAWNVFCTVLGPYSFRAVFAVFIVVALIRRHFRPAVFA-LL
Mvan_0429|M.vanbaalenii_PYR-1 FGDGRPRWVTAWDVLCTLLGPGAFRLAVLVVIVVALVRRRFRLAVFL-TV
Mflv_0312|M.gilvum_PYR-GCK VGESHSGWVTAWDVFCTVLGPGAFRLAVLVLIVLALLRRRIRLAVFL-VV
MAB_0200c|M.abscessus_ATCC_199 YGIAHPGWVTVWDVFATALGPGAFRIGGAIIIIVLLVRGQRRPAVFL-LV
TH_3242|M.thermoresistible__bu FGADRPAWVMSWAVFCTVLGPMAFRLAALIAVVAAVRRRSPRVAVFL-LA
MLBr_00094|M.leprae_Br4923 ISLLGAILVPCRRRYWLVAGAGVFAAHVAAVLIKRMVRR-IRPNHPA--V
. . : *. : *
Mb0316|M.bovis_AF2122/97 CLPLNAIMTIAAKSVAHRPRPATALVSAHSTSFPSGHALEATASVLALLT
Rv0308|M.tuberculosis_H37Rv CLPLNAIMTIAAKSVAHRPRPATALVSAHSTSFPSGHALEATASVLALLT
MMAR_0558|M.marinum_M CVPLNWIITMIAKGLADRPRPVTKLVAAHSASFPSGHALELTASVLAMLT
MUL_1222|M.ulcerans_Agy99 CVPLNWIVTMIAKGLADRPRPVTKLVAAHSASFPSGHALELTASVLAMLT
MAV_4854|M.avium_104 CAPCNELVTAGAKALVDRPRPPTALVGAASTSFPSGHALEATAGLLAMTL
MSMEG_0634|M.smegmatis_MC2_155 SVELSGLVTQLAKLASDRPRPPTALVYASWTSFPSGHALGVMVCVPALLA
Mvan_0429|M.vanbaalenii_PYR-1 TVELSAVVTETAKYIVDRPRPDTAMVSALSTSFPSGHALGVMVSVAALLT
Mflv_0312|M.gilvum_PYR-GCK TVELSGLVTEIAKHLAGRARPDTAFVDAWGLSFPSGHALGVLVAVAALLS
MAB_0200c|M.abscessus_ATCC_199 TIELSALVTELAKLLAGRPRPATKLVDAYGTSFPSGHALGIMVCVLALLT
TH_3242|M.thermoresistible__bu TVVFNGVLIEGAKWLADRPRPATALVPAWSTAFPSGHAVGVLLAVLALLT
MLBr_00094|M.leprae_Br4923 TVNVGTPSPLSFPSAHATSTAAAAILIGRASRLPKGIVAAVLVAPMALSR
. . . : :: . :*.* . *:
Mb0316|M.bovis_AF2122/97 VLLPMLHSRFTRHIAITVGALCVLTVGVARVALNVHHPTDVVAGWALGYL
Rv0308|M.tuberculosis_H37Rv VLLPMLHSRFTRHIAITVGALCVLTVGVARVALNVHHPTDVVAGWALGYL
MMAR_0558|M.marinum_M FLLPMIRVHWMRILAVVVGALSVLLVGISRVALNVHHPSDVIAGWALGYV
MUL_1222|M.ulcerans_Agy99 FLLPMIRVHWMRILAVVVGALSVLLVGISRVALNVHHPSDVIAGWALGYV
MAV_4854|M.avium_104 FALPMLN-RVLRGLLVAAVAVALPAVGLSRVMLNVHYPSDVLAGWSLGYL
MSMEG_0634|M.smegmatis_MC2_155 LVWPLLRGAWRGWA-VAAGALVIVAIGVGRVVLNVHHPSDVVAGWALGYA
Mvan_0429|M.vanbaalenii_PYR-1 VVLPVVRPALRGWL-VALGIAVILVVGIGRVVLNVHHPSDVIAGWALGYA
Mflv_0312|M.gilvum_PYR-GCK VAVPAVAPSRRAWL-IALGVVIVVTVGVGRVILNVHHPSDVLAGWALGYA
MAB_0200c|M.abscessus_ATCC_199 VFTPKVVAAWRPWL-GVLGGVLILLIGISRVLLNVHNPSDVLAGWALGYA
TH_3242|M.thermoresistible__bu VFWPKIDR-RAGWT-LAA-AVVVFAIGAGRVVLNVHHPSDVLAGWALGYA
MLBr_00094|M.leprae_Br4923 IVLGVHYPSDVAFG---------VVLGAAVAGTTARFDSRLSRRWTVQHG
. :* . . ..: : : *:: :
Mb0316|M.bovis_AF2122/97 YFLVCLCVFRPPSIFGAQRASHALSPPVEVSRQPEPEVDTAR
Rv0308|M.tuberculosis_H37Rv YFLVCLCVFRPPSIFGAQRASHALSPPVEVSRQPEPEVDTAR
MMAR_0558|M.marinum_M YFLFWLWVLR----------------PVPVAEQEPAELVSAG
MUL_1222|M.ulcerans_Agy99 YFLFWLWVLR----------------PVPVAEQEPAELVSAG
MAV_4854|M.avium_104 YFLLCLLVFR------------------------PSAPDRSG
MSMEG_0634|M.smegmatis_MC2_155 FFVVCLL--LAPPYPDVRARGE-----------IPEVRGIAT
Mvan_0429|M.vanbaalenii_PYR-1 YFVACLL--LVWPSVPVIERAE-----------KPAVLDNAH
Mflv_0312|M.gilvum_PYR-GCK YFLVCLR--LVPPAT-LTRRDE-----------TPAELDIAR
MAB_0200c|M.abscessus_ATCC_199 YFIVCLL--ALPPWT-VTAADG-----------TPPAPGNAP
TH_3242|M.thermoresistible__bu WFVGCYLGCLRPTWPDRPARAEP----VTGGARTPAGPDSAP
MLBr_00094|M.leprae_Br4923 LSSGSAVK----------------------------------