For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MELGLHFIDFLPGDPARLGPTLSEAARAAEQGGATLFTLADHFFQMEQLGPAENPFLEGYTSLGFLAAQT STIELALLVTGVTYRYPGVLAKTVTTLDVLSQGRSILGLGAAWYEREHTALGIPYPALSTRFEMLEETLQ ICLQMWSDDNGAYQGKHYRLAETICAPQPIRRPPILIGGDGEKETLRLVAQYADVWNSMSNDLAELEHKI EVLHRHCDAVGRDPRDIRKTCGGLATLDPFEDLDEYLRTVERLGRLGIEMINVGPLPGNPDPVGYIRRLG DEVIPRLAEIG
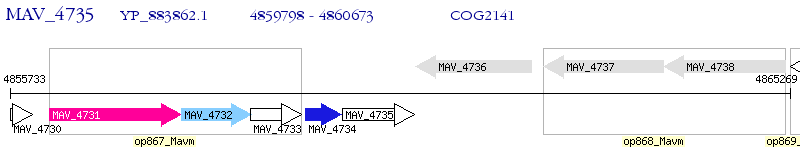
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_4735 | - | - | 100% (291) | hypothetical protein MAV_4735 |
| M. avium 104 | MAV_2865 | - | 1e-47 | 38.69% (274) | hypothetical protein MAV_2865 |
| M. avium 104 | MAV_0169 | - | 6e-36 | 37.29% (236) | hypothetical protein MAV_0169 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1886c | - | 3e-43 | 42.86% (224) | oxidoreductase |
| M. gilvum PYR-GCK | Mflv_3347 | - | 3e-43 | 40.44% (225) | luciferase family protein |
| M. tuberculosis H37Rv | Rv1855c | - | 3e-43 | 42.86% (224) | oxidoreductase |
| M. leprae Br4923 | MLBr_00269 | - | 1e-10 | 27.43% (226) | putative F420-dependent glucose-6-phosphate dehydrogenase |
| M. abscessus ATCC 19977 | MAB_2431c | - | 1e-48 | 46.41% (209) | luciferase-like oxidoreductase |
| M. marinum M | MMAR_2729 | - | 5e-44 | 43.75% (208) | oxidoreductase |
| M. smegmatis MC2 155 | MSMEG_1749 | - | 6e-81 | 52.53% (297) | putative monooxygenase |
| M. thermoresistible (build 8) | TH_1174 | - | 2e-44 | 41.33% (225) | POSSIBLE OXIDOREDUCTASE |
| M. ulcerans Agy99 | MUL_3024 | - | 9e-44 | 43.27% (208) | oxidoreductase |
| M. vanbaalenii PYR-1 | Mvan_3077 | - | 1e-46 | 42.22% (225) | luciferase family protein |
CLUSTAL 2.0.9 multiple sequence alignment
MAV_4735|M.avium_104 ---MELGLHFIDF-LPGDPARLGPTLSEAARAAEQGGATLFTLADHFFQM
MSMEG_1749|M.smegmatis_MC2_155 ---MDLGFHIPIFDIDGGTTAIAGELARVGESAEAAGASWLSFMDHFFQI
Mb1886c|M.bovis_AF2122/97 -MTIRLGLQIPNFSYGTGVEKLFPSVIAQAREAEAAGYDSLFVMDHFYQL
Rv1855c|M.tuberculosis_H37Rv -MTIRLGLQIPNFSYGTGVEKLFPSVIAQAREAEAAGYDSLFVMDHFYQL
MMAR_2729|M.marinum_M -MAIRLGFQIPSFSYGTGVDKLFPSVIAQAREAEAAGFDSLFLMDHFYQL
MUL_3024|M.ulcerans_Agy99 -MAIRLGFQIPSFSYGTGVDKLFPSVIAQAREAEAAGFDSLFLMDHFYQL
Mflv_3347|M.gilvum_PYR-GCK -MSIRLGLQINNFSYGTGVSDLFPTVVAQAKEADTSGFDSVFLMDHFYQL
Mvan_3077|M.vanbaalenii_PYR-1 -MTIRLGLQINNFSYGTPVPDLFPTVVAQAQEADASGFDSVFVMDHFYQL
TH_1174|M.thermoresistible__bu -VSIRLGLQIPNFSYGTGVADLFPTVIAQAREADEAGYDAVFVMDHFYQL
MAB_2431c|M.abscessus_ATCC_199 -MTIRLGYQIPNFSYGTGVAELFPTVVKQVREAEASGFDTVFVMDHFYQL
MLBr_00269|M.leprae_Br4923 MAELRLGYKASAEQFAPR------ELVELGVAAEAHGMDSATVSDHFQPW
: ** : : *: * . ***
MAV_4735|M.avium_104 EQLG-PAENPFLEGYTSLGFLAAQTSTIELALLVTGVTYRY-PGVLAKTV
MSMEG_1749|M.smegmatis_MC2_155 EPTGLPAEAHMLEGYTTLGYLAAHTETVPLGLLVTGVTYRH-PGLLAKIV
Mb1886c|M.bovis_AF2122/97 PMLG-TPDQPMLEAYTALGALATATERLQLGALVTGNTYRS-PTLLAKII
Rv1855c|M.tuberculosis_H37Rv PMLG-TPDQPMLEAYTALGALATATERLQLGALVTGNTYRS-PTLLAKII
MMAR_2729|M.marinum_M PMLG-TPDQPMLEAYTALGALATATERLQLGTLVTGNTYRN-PALLAKII
MUL_3024|M.ulcerans_Agy99 PMLG-TPDQPMLEAYTALGALATATERLQLGTLVTGNTYRN-PALLAKII
Mflv_3347|M.gilvum_PYR-GCK PGIG-TPDQPMLEAYTALGALAAATDSVQLGTLVTGNTYRN-PTLLAKNI
Mvan_3077|M.vanbaalenii_PYR-1 PGLG-TPDQPMLEAYTALGALAAVTQNVQLGTLVTGNTYRN-PTLLAKNI
TH_1174|M.thermoresistible__bu PMLG-DPDEPMLEAYTALGGLATSTERVQLGTLVTGNTYRN-PTLLAKAV
MAB_2431c|M.abscessus_ATCC_199 PMIG-TPDQPMLEAYTALGALASATSTIQLSTLVTGNTYRN-PALLAKAV
MLBr_00269|M.leprae_Br4923 RHQG----GHASFSLSWMTAVGERTNRILLGTSVLTPTFRYNPAVIGQAF
* . : : :. *. : *. * *:* * ::.: .
MAV_4735|M.avium_104 TTLDVLSQGRSILGLGAAWYEREHTALGIP--YPALSTRFEMLEETLQIC
MSMEG_1749|M.smegmatis_MC2_155 TTLDVLSGGRAVLGVGAAWFEREHVGLGVP--FPPLAERFERLEETLQIC
Mb1886c|M.bovis_AF2122/97 TTLDVVSAGRAILGIGAGWFELEHRQLGFE--FGTFSDRFNRLEEALQIL
Rv1855c|M.tuberculosis_H37Rv TTLDVVSAGRAILGIGAGWFELEHRQLGFE--FGTFSDRFNRLEEALQIL
MMAR_2729|M.marinum_M TTLDVVSAGRAVLGIGAGWFELEHRQLGYE--FGTFTDRFNRLEEALQIL
MUL_3024|M.ulcerans_Agy99 TTLDVVSAGRAVLGIGAGWFELEHRQLGYE--FGTFTDRFNRLEEALQIL
Mflv_3347|M.gilvum_PYR-GCK TTLDVISAGRAILGIGTGWFELEHDQLGYE--FGTFTDRFNKLGEALQII
Mvan_3077|M.vanbaalenii_PYR-1 TTLDVISQGRAVLGIGTGWFELEHDQLGYE--FGTFTERFDKLDEALQII
TH_1174|M.thermoresistible__bu TTLDVMSQGRAVLGIGTGWFELEHQQLGYE--FGTFTDRFNRLYEALEII
MAB_2431c|M.abscessus_ATCC_199 TTLDVVSGGRAILGIGAGWFELEHQQLGFE--FGTFTDRFERLTEALEII
MLBr_00269|M.leprae_Br4923 ATMGCLYPNRVFLGVGTG-EALNEVATGYQGAWPEFKERFARLRESVRLM
:*:. : .* .**:*:. :. * : : ** * *::.:
MAV_4735|M.avium_104 LQMWS-DDNGAYQGKHYRLAETICAPQPIRRPPILIGGDGEKETLRLVAQ
MSMEG_1749|M.smegmatis_MC2_155 AQMWDPADNGPYDGKYYQLAETLCSPQPINRPKVMIGGSGERKTLRLVAQ
Mb1886c|M.bovis_AF2122/97 EPMVK-GERPTFFGDWYTTESAMAEPRYRDRIPILIGGGGEKKTFAIAAR
Rv1855c|M.tuberculosis_H37Rv EPMVK-GERPTFFGDWYTTESAMAEPRYRDRIPILIGGGGEKKTFAIAAR
MMAR_2729|M.marinum_M DPMIK-GERPTFSGSWYQAESAMAEPRYRDRIPILIGGGGEKKTFRIAAH
MUL_3024|M.ulcerans_Agy99 DPIIK-GERPTFSGSWYQAESAMAEPRYRDRIPILIGGGGEKKTFRIAAH
Mflv_3347|M.gilvum_PYR-GCK LPMIK-GETPTFSGEYYSVKDAMANPRFREHIPLMIGGSGEKKTIPLAAR
Mvan_3077|M.vanbaalenii_PYR-1 VPMLK-GERPTFTGRHYRVQEAMANPRFREHIPLMIGGSGEKKTIPLAAR
TH_1174|M.thermoresistible__bu VPMIE-GKRPTFEGRYYRVESAAAEPRFRDRIPLMIGGSGEKKTIPLAAR
MAB_2431c|M.abscessus_ATCC_199 PPMLR-GERPTFEGKYYRTESALNEPRLRPNIPIMIGGSGEKKTFGLAVK
MLBr_00269|M.leprae_Br4923 RELWR-GDRVDFDGDYYQLKGASIYDVPEGGVPIYIAAGGP-EVAKYAGR
: . : * * : : *...* :. . :
MAV_4735|M.avium_104 YADVWNSMSNDLAELE--HKIEVLHRHCDAVGRDPRDIRKTCGGLATLDP
MSMEG_1749|M.smegmatis_MC2_155 YGDACNLFASSVDEVR--HKIDVLRRHCDTVGRDFGEIRVTI--IANNDP
Mb1886c|M.bovis_AF2122/97 FADHLNIVAA-VDELP--RKMRALAARCDEAGRDRSTLQTSLLLTVMIDE
Rv1855c|M.tuberculosis_H37Rv FADHLNIVAA-VDELP--RKMRALAARCDEAGRDRSTLQTSLLLTVMIDE
MMAR_2729|M.marinum_M WADHLNIVAA-LDELP--RKMDAVAARCEEAGRDPSTLETSVMLTVVVDE
MUL_3024|M.ulcerans_Agy99 WADHLNIVAA-LDELP--RKMDAVAARCEEAGRDPSTLETSVMLTVVVDA
Mflv_3347|M.gilvum_PYR-GCK HFDHLNVIAG-FDELA--RKIDVVTQACEKIDRDPSTLETSMLIGALTGD
Mvan_3077|M.vanbaalenii_PYR-1 HFDHLNVIAG-FDELA--RKIEVIEQRCADIDRDPATLETSVLVGALVGD
TH_1174|M.thermoresistible__bu RFDHLNLIAG-FDELP--GKVAAVKRSCEEIGRDPDTLETSTLVSVMVDD
MAB_2431c|M.abscessus_ATCC_199 YADHLNLLSS-LDEIP--HKLTVLRQRCEEAGRDPSTLETSAFFSVFIDD
MLBr_00269|M.leprae_Br4923 AGEGFVCTSGKGEELYTEKLIPAVLEGAAVAGRDADDIDKMIEIKMSYDP
: : *: : .: . .** : .
MAV_4735|M.avium_104 ------------------------------FEDLDEYLR-----------
MSMEG_1749|M.smegmatis_MC2_155 R---------------------------PTPETRDEFVR-----------
Mb1886c|M.bovis_AF2122/97 T---------------------------LSPDAIPAEMS-----GRVVVG
Rv1855c|M.tuberculosis_H37Rv T---------------------------LSPDAIPAEMS-----GRVVVG
MMAR_2729|M.marinum_M K---------------------------AKVERLPAETS-----RRMVTG
MUL_3024|M.ulcerans_Agy99 K---------------------------AKVERLPAETS-----RRMVTG
Mflv_3347|M.gilvum_PYR-GCK G---------------------------VSPDMIPEDFR-----QRMVAG
Mvan_3077|M.vanbaalenii_PYR-1 D---------------------------VSPDQIPDEFK-----QRMVAG
TH_1174|M.thermoresistible__bu N---------------------------ITDDMLPEKAR-----GRLLVG
MAB_2431c|M.abscessus_ATCC_199 DSATAWK-----------LAASQLSQNGIDLDTLPDEARAQ-VLGRTLVG
MLBr_00269|M.leprae_Br4923 DPEQALSNIRFWAPLSLAAEQKHSIDDPIEMEKVADALPIEQVAKRWIVV
:
MAV_4735|M.avium_104 --------TVERLGRLGIEMINVGPLPGN-PDPVGYIRRLGDEVIPRLAE
MSMEG_1749|M.smegmatis_MC2_155 --------SMADYAKLGVHTTIVIPTTG---SPAAWIDGM-APAVPQLAE
Mb1886c|M.bovis_AF2122/97 SPAQIADQIQAKVLDAGVDGLIINLAPH--GYLPGVITTAAEALRPLLGV
Rv1855c|M.tuberculosis_H37Rv SPAQIADQIQAKVLDAGVDGLIINLAPH--GYLPGVITTAAEALRPLLGV
MMAR_2729|M.marinum_M SPAQIADQVQTKVLDTGIDGVIMNLSAH--GHSPGLITTVAEALRPLLTG
MUL_3024|M.ulcerans_Agy99 SPAQIADQVQTKVLDTGIDGVIMNLSAH--GHSPGLITTVAEALRPLLTG
Mflv_3347|M.gilvum_PYR-GCK TPEQIAEQVQSKVLDAGVDGVILFVPTQTVGYQPGQITALGEALKPLVAG
Mvan_3077|M.vanbaalenii_PYR-1 TVDQVAEQIKTKVLDAGIDGVILYVPTQIVGYQPGQITALGEALAPLVTA
TH_1174|M.thermoresistible__bu SAERIAEQLQTKVLDAGVDGVIVNMATKVQGYRPGLITALGEALRPRVGA
MAB_2431c|M.abscessus_ATCC_199 NPEEIAEQVQTRVLDQGIDSVTLNAPAT--GHLPGVVELIGNTLRPLFKN
MLBr_00269|M.leprae_Br4923 SDPDEAVARVGQYVTWGLNHLVFHAPGH---NQRRFLELFEKDLAPRLRR
*:. . : * .
MAV_4735|M.avium_104 IG-
MSMEG_1749|M.smegmatis_MC2_155 LGS
Mb1886c|M.bovis_AF2122/97 ---
Rv1855c|M.tuberculosis_H37Rv ---
MMAR_2729|M.marinum_M ---
MUL_3024|M.ulcerans_Agy99 ---
Mflv_3347|M.gilvum_PYR-GCK ---
Mvan_3077|M.vanbaalenii_PYR-1 ---
TH_1174|M.thermoresistible__bu ---
MAB_2431c|M.abscessus_ATCC_199 ---
MLBr_00269|M.leprae_Br4923 LG-