For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VTGSGSARQLTLTARLNTSAVDSRRGVVRLHPSAVAALGIREWDAVSLIGSRTTAAVAGLAGPEIPSGTV LLDDVTLSNAGLRDGTAVVVSPVTVYGARSVTLSGSPLTTRSVSPVTLRQALLGKVLTVGDAVSLLPRDL GPGTSTSEATRALAAAVGISWTSELLTVTGVDPDGPVSVQPNSLVGWGARAAPAAASRVSVEVARPQTVV EELKGCQPQAAKLVEWLKLALDEPHLLKTLGAATFLGVLVTGPAGVGKVTLVRAVCGDRRLVTLDGPEVG ALGADDRLKAVASAVQTVRDGGGVLLITDVDALLPATAEPVASLILAELRGAVASEGVALIATSARPDQL DARLRAPDLCDRELSLPLPDAATRKALLESLLRNVPTDGLDLDEIASCTPGFVVADLAALIREAALRAAS RASSDGRPPALHQEDLTGALSVIRPLSRSAAEEVSVGDITLDDVGDMAQARQALTEAVLWPLQHPDTFAR LGVQPPRGVLLYGPPGCGKTFVVRALASTGQLSVHAVKGSELMDKWVGSSEKAVRELFRRARDSAPALIF LDEVDALAPRRGQSFDSGVTDRVVAALLTELDGVDPLRDVVVLGATNRPDLIDPALLRPGRLERLVFVEP PDAAARREILRTAGKSIPLSADVDLDELAAGLDGYSAADCVALLREAALTAMRRSIDATDVTAADLAAAR ENVRPSLDPAQVASLRAFAEAP
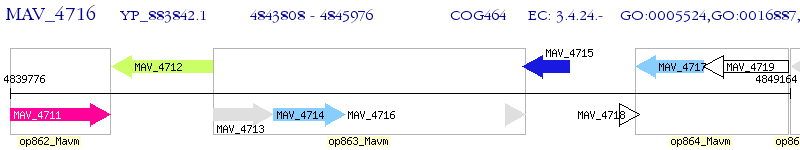
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_4716 | - | - | 100% (722) | cell division protein 48 |
| M. avium 104 | MAV_0542 | hflB | 9e-46 | 39.62% (260) | ATP-dependent metallopeptidase HflB |
| M. avium 104 | MAV_2400 | - | 3e-42 | 37.70% (252) | ATPase, AAA family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0443c | - | 0.0 | 85.42% (720) | putative ATPase |
| M. gilvum PYR-GCK | Mflv_0155 | - | 0.0 | 76.03% (726) | vesicle-fusing ATPase |
| M. tuberculosis H37Rv | Rv0435c | - | 0.0 | 85.42% (720) | putative ATPase |
| M. leprae Br4923 | MLBr_01316 | - | 2e-44 | 34.55% (356) | putative AAA-family ATPase |
| M. abscessus ATCC 19977 | MAB_0637c | - | 0.0 | 75.80% (723) | putative ATPase |
| M. marinum M | MMAR_0752 | - | 0.0 | 85.18% (722) | ATPase |
| M. smegmatis MC2 155 | MSMEG_0858 | - | 0.0 | 75.62% (722) | cell division control protein Cdc48 |
| M. thermoresistible (build 8) | TH_2865 | - | 0.0 | 75.68% (740) | PUTATIVE Vesicle-fusing ATPase |
| M. ulcerans Agy99 | MUL_1384 | - | 0.0 | 84.92% (723) | ATPase |
| M. vanbaalenii PYR-1 | Mvan_0749 | - | 0.0 | 76.00% (725) | vesicle-fusing ATPase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb0443c|M.bovis_AF2122/97 ------------MTHPDPARQLTLTARLNTSAVDSRRGVVRLHPNAIAAL
Rv0435c|M.tuberculosis_H37Rv ------------MTHPDPARQLTLTARLNTSAVDSRRGVVRLHPNAIAAL
MMAR_0752|M.marinum_M ------------MTSPYGAPQLTLTARLNTSAVDSRRGVVRLHPSAIAAL
MUL_1384|M.ulcerans_Agy99 ------------MTSPYGAPQLTLTARLNTSAVDSRRGVVRLHPSAIAAL
MAV_4716|M.avium_104 ------------MTGSGSARQLTLTARLNTSAVDSRRGVVRLHPSAVAAL
Mflv_0155|M.gilvum_PYR-GCK ------------------MAQLTLTARLNTSALDSRRGVVRLHPEAIAAL
Mvan_0749|M.vanbaalenii_PYR-1 -----MSELHQSAEHRPPRAHLTLTARLNTSALDSRRGVVRLHPEAIAAL
TH_2865|M.thermoresistible__bu -------VTVTTDVPEPELNQLRLTARLNTSALDSRRGVVRLHPEAIAAL
MAB_0637c|M.abscessus_ATCC_199 ------------------MSSLTLTARLNTSAVDSRRGVVRLHPEAIAAL
MSMEG_0858|M.smegmatis_MC2_155 MSQGFDDEFGGADQYDTPLSHLRLTARLNTSALDSRRGVVRLHPEVLAAL
MLBr_01316|M.leprae_Br4923 -----------------------MGESERSEAFNPPR-------------
: .:.*.:. *
Mb0443c|M.bovis_AF2122/97 GIREWDAVSLTGSRTTAAVAGLAAADTAVGTVLLDDVTLSNAGLREGTEV
Rv0435c|M.tuberculosis_H37Rv GIREWDAVSLTGSRTTAAVAGLAAADTAVGTVLLDDVTLSNAGLREGTEV
MMAR_0752|M.marinum_M GIREWDAVSLTGSRTTAAVAGMADDQVPVGTILLDDVTLSNAGLREGTAV
MUL_1384|M.ulcerans_Agy99 GIREWDAVSLTGSRTTAAVAGMADDQVPVGTILLDDVTLSNAGLREGTAV
MAV_4716|M.avium_104 GIREWDAVSLIGSRTTAAVAGLAGPEIPSGTVLLDDVTLSNAGLRDGTAV
Mflv_0155|M.gilvum_PYR-GCK GIREWDAVSLTGARTTAAVVGIAPAGTPTGTALLDDVTLSNAGLREDTSV
Mvan_0749|M.vanbaalenii_PYR-1 GIREWDAVSLTGARTTAAVVGIAPPGTPTGTALLDDVTLSNAGLREDTSV
TH_2865|M.thermoresistible__bu GLREWDAVSLTGSRTTAAVVGHAPPGTPVGTALLDDVTLSNAGLVEDAPV
MAB_0637c|M.abscessus_ATCC_199 GIREWDAISLTGSRTTAAVVGVAPRSVPAGTALLDDVTLSNAGLREDASV
MSMEG_0858|M.smegmatis_MC2_155 GIREWDAVALTGTRTTAAVAGVAGPGVPAGTALLDDVTLSNAGVRENAAV
MLBr_01316|M.leprae_Br4923 ------------------EAGMSSGDIAELEQLRREIVVLREQLEHAVGP
.* : . * ::.: . : . .
Mb0443c|M.bovis_AF2122/97 IVSPVTVYGARSVTLSGSTLATQSVPPVTLRQALLGKVMTVGDAVSLLPR
Rv0435c|M.tuberculosis_H37Rv IVSPVTVYGARSVTLSGSTLATQSVPPVTLRQALLGKVMTVGDAVSLLPR
MMAR_0752|M.marinum_M LVNSVTVYGARTVTLSGSSLATQSLSPITLRQALLGKVMTVGDAVSLLPR
MUL_1384|M.ulcerans_Agy99 LVNSVTVYGARTVTLSGSSLATQSLSPITLRQALLGKVMTVGDAVSLLPR
MAV_4716|M.avium_104 VVSPVTVYGARSVTLSGSPLTTRSVSPVTLRQALLGKVLTVGDAVSLLPR
Mflv_0155|M.gilvum_PYR-GCK LVSDVTVYGARSVTLSGSRLASRSITPATLRQALLGKVMTVGDTVSLLPR
Mvan_0749|M.vanbaalenii_PYR-1 LVAPVTVYGARCVTLSGSKLAMQSITAATLRQALLGKVMTVGDTVSLLPR
TH_2865|M.thermoresistible__bu LVAPVTVYGARSVTVAGSRLASGSISPATLRQALLGKVMTVGDTVSLLPR
MAB_0637c|M.abscessus_ATCC_199 IVAAVTVYGARSITVSGSSIATQSVSSATLRMALLGKVMTVGDTVSLLPR
MSMEG_0858|M.smegmatis_MC2_155 LVSPVTVYGARSVTVSGSRLATQSISPATLRMALLGKVMTVGDTVSLLPR
MLBr_01316|M.leprae_Br4923 HGSVRSVRDVHQLEARIDSLTARNSKLMDTLKEARQQLLALREEVDRLGQ
:* ..: : . :: . ::::: : *. * :
Mb0443c|M.bovis_AF2122/97 DLGPGTSTSAASRALAAAVGISWTSELLTVTGVDPDGPVSVQPNSLVTWG
Rv0435c|M.tuberculosis_H37Rv DLGPGTSTSAASRALAAAVGISWTSELLTVTGVDPDGPVSVQPNSLVTWG
MMAR_0752|M.marinum_M DLGPGTSTSAASQALASAVGISWTSELLTVTGVDPDAPVSVQPNTLVTWG
MUL_1384|M.ulcerans_Agy99 DLGPGTSTSAASQALASAVGISWTSELLTVTGVDPDAPVSVQPNTLVTWG
MAV_4716|M.avium_104 DLGPGTSTSEATRALAAAVGISWTSELLTVTGVDPDGPVSVQPNSLVGWG
Mflv_0155|M.gilvum_PYR-GCK DLGPGTSTSTASAALASSVGITWTSELLTVTAADPIGPVSVQPNSLVCWA
Mvan_0749|M.vanbaalenii_PYR-1 DLGPGTSTSAASAALASSVGITWTSELLTVTSVDPAGPVSVQPNSLVNWG
TH_2865|M.thermoresistible__bu DLGPGTSTSAATTALAASVGITWTSELLTVTAVDPPGPVSVQPNSVVAWA
MAB_0637c|M.abscessus_ATCC_199 DLGPGTSTSEATSALTRSVGITWTSELLTVTAVDPDGPVSVQPNSLVSWQ
MSMEG_0858|M.smegmatis_MC2_155 DLGPGTSTSAATSALASSVGITWTSELLTVTAVDPPGTVSVQPNSVVSWG
MLBr_01316|M.leprae_Br4923 PPSG-----------YGVLLAAHDDETVDVFTSGRKMRLTCSPN------
. : : .* : * . :: .**
Mb0443c|M.bovis_AF2122/97 AGVPAAMGTSTAGQVSISSP--------------EIQIEELKGAQPQAAK
Rv0435c|M.tuberculosis_H37Rv AGVPAAMGTSTAGQVSISSP--------------EIQIEELKGAQPQAAK
MMAR_0752|M.marinum_M TGVPHTAQSSQAERVSVATP--------------EIQVEELKGAQPQAAK
MUL_1384|M.ulcerans_Agy99 TGVPHTAQSSQAERVSVATP--------------EIQVEELKGAQPQAAK
MAV_4716|M.avium_104 ARAAPAAASRVS--VEVARP--------------QTVVEELKGCQPQAAK
Mflv_0155|M.gilvum_PYR-GCK SGASEASGTSEAANGTSEVSGAPVRTEQP--RTQAVGFDDLKGAHAQAGR
Mvan_0749|M.vanbaalenii_PYR-1 TSTATPAAPPEGT-GQHVVT----TSAES--TTPAVSYDDLKGSHAQAAR
TH_2865|M.thermoresistible__bu DSTGTHGTGTHGTGTHGTGTAAPARPPGPGRARPDFTVEDLKGAQSQAER
MAB_0637c|M.abscessus_ATCC_199 GSTATSATSATAARQTELAPAAGPAPAAS----TVISVEDLKGSQAQATR
MSMEG_0858|M.smegmatis_MC2_155 TGTPEDPAPPPTGRHTVSPQ----RSEQP------VSFDDVKVTHPQAVK
MLBr_01316|M.leprae_Br4923 -----------------------------------IEVASLR--KGQTVR
.:: : *: :
Mb0443c|M.bovis_AF2122/97 LTEWLKLALDEPHLLQTLGAGTNLGVLVSGPAGVGKATLVRAVCDGRRLV
Rv0435c|M.tuberculosis_H37Rv LTEWLKLALDEPHLLQTLGAGTNLGVLVSGPAGVGKATLVRAVCDGRRLV
MMAR_0752|M.marinum_M LTEWLKLALDEPHLLQSLGAGTNLGVLVSGPAGVGKVTLVRAVCAGRRLV
MUL_1384|M.ulcerans_Agy99 LTEWLKLALDEPHLLQSLGAGTNLGVLASGPAGVGKVTLVRAVCAGRRLV
MAV_4716|M.avium_104 LVEWLKLALDEPHLLKTLGAATFLGVLVTGPAGVGKVTLVRAVCGDRRLV
Mflv_0155|M.gilvum_PYR-GCK LTEWLKLALDQPELLEKLGATANLGVLVSGPAGVGKATLVRTVCAERRLI
Mvan_0749|M.vanbaalenii_PYR-1 LTEWLKLAIDQPELLEKLGATSNLGVLVSGPAGVGKATLVRTVCAGRRLI
TH_2865|M.thermoresistible__bu LTEWLKLALDEPELLETLGATPHLGVLVSGPHGVGKATLVRVVCAHRRLV
MAB_0637c|M.abscessus_ATCC_199 LTEWLKLALDEPELLKTLGAQPNLGVLISGPAGVGKATLARAVCAARTLI
MSMEG_0858|M.smegmatis_MC2_155 LDEWLRLSLDEPELLKTLGATPHLGVLVSGPAGVGKATMVRAVCASRRVV
MLBr_01316|M.leprae_Br4923 LNEALTVVEAG--TFEAVGEVSTLREVLAD----GHRALVVGHADEERIV
* * * : :: :* . * : :. *: ::. . . ::
Mb0443c|M.bovis_AF2122/97 TLDGPEIGALAAGDRVKAVASAVQAVRHEGGVLLITDADALLP-----AA
Rv0435c|M.tuberculosis_H37Rv TLDGPEIGALAAGDRVKAVASAVQAVRHEGGVLLITDADALLP-----AA
MMAR_0752|M.marinum_M ELDGPEVGALAPENRLKAVASAVATVRDGGGVLLITDVDALLP-----AT
MUL_1384|M.ulcerans_Agy99 ELDGPEVGALAPENRLKAVASAVATVRDGGGVLLITDVDALLP-----AT
MAV_4716|M.avium_104 TLDGPEVGALGADDRLKAVASAVQTVRDGGGVLLITDVDALLP-----AT
Mflv_0155|M.gilvum_PYR-GCK ELDGPEVGSLRADDRLAAVSSAAASARDGGGVLLVTDIDALLP-----VP
Mvan_0749|M.vanbaalenii_PYR-1 ELDGPEVGSLRPEDRLANVSSAVAAVRDGGGVLLVTDVDALLP-----VP
TH_2865|M.thermoresistible__bu ELDGPEIGALPAEERLRRVSETVATVRDGGGVLLITDVDALLPQ-SADRP
MAB_0637c|M.abscessus_ATCC_199 ELDGPEAGATSADGRLDMIREAVSRVRQSGGVLLISDIDALLP-----AA
MSMEG_0858|M.smegmatis_MC2_155 ELDGPEVGALQVDERLRSVTSAVAAVTESGGVLFIADVDALLPAGNEMRP
MLBr_01316|M.leprae_Br4923 CLAEPLVAENLLDGVPGALNDDSRPRKLRPGDSLLVDPKAGYAFER--VP
* * . : . * :: * .* . .
Mb0443c|M.bovis_AF2122/97 AEPVASLILSELRTAVATAGVVLIATSARPDQLDARLRSPELCDRELGLP
Rv0435c|M.tuberculosis_H37Rv AEPVASLILSELRTAVATAGVVLIATSARPDQLDARLRSPELCDRELGLP
MMAR_0752|M.marinum_M AEPVAALILGELRTAVATDGVVLIATTAAPDQLDARLRAPDLCDRELGLP
MUL_1384|M.ulcerans_Agy99 AEPVAALILGELRTAVATDGVVLIATTAAPDQLDARLRAPDLCDRELGLP
MAV_4716|M.avium_104 AEPVASLILAELRGAVASEGVALIATSARPDQLDARLRAPDLCDRELSLP
Mflv_0155|M.gilvum_PYR-GCK PDPVATLILTELRSAVAG-GVVFVATSAVPDNVDPRLRAPDLCDRELGLS
Mvan_0749|M.vanbaalenii_PYR-1 AEPVATLILTELRSAVATRGVAFVATTAVPDNVDPRLRAPDLCDRELGLT
TH_2865|M.thermoresistible__bu AEPVATAILAELRDAVATRGVAFIATSAVPDAVDTRLRAPEVCDRELGLP
MAB_0637c|M.abscessus_ATCC_199 AEPVATLILAELRTAVATSGVAFIATTAHPDALDARLRGPELCDRELGLS
MSMEG_0858|M.smegmatis_MC2_155 PEPVATLILAELRKAVATPGVAFIATSAVPENVDARLRAPEVCDRELGLS
MLBr_01316|M.leprae_Br4923 KAEVEDLVLEEVP------DVSYQDIGGLTRQIEQIRDAVELP--FLHKE
* :* *: .* . . :: . :: *
Mb0443c|M.bovis_AF2122/97 LPDAATRKSLLEALLNPVPTGDLNLDEIASRTPGFVVADLAALVREAALR
Rv0435c|M.tuberculosis_H37Rv LPDAATRKSLLEALLNPVPTGDLNLDEIASRTPGFVVADLAALVREAALR
MMAR_0752|M.marinum_M LPDAATRKALLEALLRHVPTGDLDIDEIASRTPGFVVADLAALLREAALR
MUL_1384|M.ulcerans_Agy99 LPDAATRKALLEALLRHVPTGDLDIDEIASRTPGFVVADLAALLREAALR
MAV_4716|M.avium_104 LPDAATRKALLESLLRNVPTDGLDLDEIASCTPGFVVADLAALIREAALR
Mflv_0155|M.gilvum_PYR-GCK LPDGAVRKQLLEVLLRDVPSANLEIGEITDRTPGFVVADLAALVREAALR
Mvan_0749|M.vanbaalenii_PYR-1 LPDGAVRAQLLEVLLRDVPADDLEISEIADRTPGFVVADLAALVREAALR
TH_2865|M.thermoresistible__bu LPDAAIREQQLAVLLRNVPSEDLRLSEIADRTPGFVIADLQALIREAALR
MAB_0637c|M.abscessus_ATCC_199 LPDAATRKSLLEVLLAKVPSQNLQLDEIASRTPGFVVADLAALVREAALR
MSMEG_0858|M.smegmatis_MC2_155 LPDATARRSLLEMLLRGVPSEDLDLGDIADHTPGFVVADLAAVVREGALR
MLBr_01316|M.leprae_Br4923 LYREYALRPPKGVLLYGPPGCGKTLIAKAVANS---LAKKMAEVRGDDAR
* ** * . : : .. :*. * :* *
Mb0443c|M.bovis_AF2122/97 AASRASADGRPPMLHQDDLLGALTVIRPLSRSASDEVTVGD---VTLDDV
Rv0435c|M.tuberculosis_H37Rv AASRASADGRPPMLHQDDLLGALTVIRPLSRSASDEVTVGD---VTLDDV
MMAR_0752|M.marinum_M AASRASTDGQPPELNQEDLLGALTVIRPLSRSASSEVAVGS---ITLDDV
MUL_1384|M.ulcerans_Agy99 AASRASTDGQPPELNQEDLLGALTVIRPLSRSASSEVAVGS---ITLDDV
MAV_4716|M.avium_104 AASRASSDGRPPALHQEDLTGALSVIRPLSRSAAEEVSVGD---ITLDDV
Mflv_0155|M.gilvum_PYR-GCK AAARASENGEPPALRQEDLVGALGVIRPLSRSATEEVSVGA---VTLDDV
Mvan_0749|M.vanbaalenii_PYR-1 AAARASENGEKPALRQEDLIGALGVIRPLSRSATEEVSVGS---VTLDDV
TH_2865|M.thermoresistible__bu AAARASATGAAPVLRQQDLLGALSVIRPLSRSATEEVAVGS---VTFDDV
MAB_0637c|M.abscessus_ATCC_199 AAARASEKASAPALAQEDLIGALSVIRPLSRSTTEEVAVGS---ITLDDV
MSMEG_0858|M.smegmatis_MC2_155 AAARASSSDDDPVLRHADLEGALTVIRPLSRSASEEVSVGS---VTLDDV
MLBr_01316|M.leprae_Br4923 EAKSYFLNIKGPELLNKFVGETERHIRLIFQRAREKASEGTPVIVFFDEM
* * * : : : ** : : : .:.: * : :*::
Mb0443c|M.bovis_AF2122/97 GDMAAAKQALTEAVLWPLQHPDTFARLG-VEPPRGVLLYGPPGCGKTFVV
Rv0435c|M.tuberculosis_H37Rv GDMAAAKQALTEAVLWPLQHPDTFARLG-VEPPRGVLLYGPPGCGKTFVV
MMAR_0752|M.marinum_M GDMAQAKQALTEAVLWPLQHPDTFARLG-VEPPRGVLLYGPPGCGKTFVV
MUL_1384|M.ulcerans_Agy99 GDMAQAKQALTEAVLWPLQHPDTFARLG-VEPPRGVLLYGPPGCGKTFVV
MAV_4716|M.avium_104 GDMAQARQALTEAVLWPLQHPDTFARLG-VQPPRGVLLYGPPGCGKTFVV
Mflv_0155|M.gilvum_PYR-GCK GDMAETKQALTEAVLWPLQHPETFQRLG-VEPPRGVLLYGPPGCGKTFVV
Mvan_0749|M.vanbaalenii_PYR-1 GDMATTKQALTEAVLWPLQHPETFQRLG-VQPPRGVLLYGPPGCGKTFVV
TH_2865|M.thermoresistible__bu GDMAETKQALTEAVLWPLQHPDTFARLG-VQPPRGVLLYGPPGCGKTFVV
MAB_0637c|M.abscessus_ATCC_199 GDMVATKQALTEAVLWPLQHPDTFQRLG-VDPPRGVLLYGPPGCGKTFVV
MSMEG_0858|M.smegmatis_MC2_155 GDMVETKRALTEAVLWPLQHPDTFSRLG-IDPPRGVLLYGPPGCGKTFVV
MLBr_01316|M.leprae_Br4923 DSIFRTRGTGVSSDVETTVVPQLLSEIDGVEGLENVIVIGASNR-EDMID
..: :: : ..: : . *: : .:. :: ..*:: *... : ::
Mb0443c|M.bovis_AF2122/97 RALASTGQLSVHAVKGSELMDKWVGSSEKAVRELFRRARDSAPSLVFLDE
Rv0435c|M.tuberculosis_H37Rv RALASTGQLSVHAVKGSELMDKWVGSSEKAVRELFRRARDSAPSLVFLDE
MMAR_0752|M.marinum_M RALASTGQLSVHAVKGSELMDKWVGSSEKAVRELFRRARDSAPSLVFLDE
MUL_1384|M.ulcerans_Agy99 RALASTGQLSVHAVKGSELMDKWVGSSEKAVRELFRRARDSAPSLVFLDE
MAV_4716|M.avium_104 RALASTGQLSVHAVKGSELMDKWVGSSEKAVRELFRRARDSAPALIFLDE
Mflv_0155|M.gilvum_PYR-GCK RALASSGRLSVHAVKGAELMDKWVGSSEKAVRELFQRARDSAPSLVFLDE
Mvan_0749|M.vanbaalenii_PYR-1 RALASSGRLSVHAVKGAELMDKWVGSSEKAVRELFRRARDSAPSLVFLDE
TH_2865|M.thermoresistible__bu RALASTGRLSVHAVKGAELMDKWVGSSEKAVRELFQRARASAPSLVFLDE
MAB_0637c|M.abscessus_ATCC_199 RALASSGRLSVHAVKGAELMDKWVGSSEQAVRDLFRRARDSAPSLVFLDE
MSMEG_0858|M.smegmatis_MC2_155 RALASSGRLSVHAVKGSELMDKWVGSSEKAVRELFARARDSAPSLVFLDE
MLBr_01316|M.leprae_Br4923 PAILRPGRLDVKIKIER--------PDAEAAQDIYSKYLTES-LPVHADD
*: .*:*.*: .. :*.:::: : .: :. *:
Mb0443c|M.bovis_AF2122/97 LDALAPRRGQSFDSGVSDRVVAALLTELDGIDPLRDVVMLGATNRPDLID
Rv0435c|M.tuberculosis_H37Rv LDALAPRRGQSFDSGVSDRVVAALLTELDGIDPLRDVVMLGATNRPDLID
MMAR_0752|M.marinum_M VDALAPRRGQSFDSGVGDRVVAALLTELDGIEPLRDVVVLGATNRPDLID
MUL_1384|M.ulcerans_Agy99 VDALAPRRGQSFDSGVGDRVVAALLTELDGIEPLRDVVVLGATNRPDLID
MAV_4716|M.avium_104 VDALAPRRGQSFDSGVTDRVVAALLTELDGVDPLRDVVVLGATNRPDLID
Mflv_0155|M.gilvum_PYR-GCK IDALAPRRGQSFDSGVTDRVVAAMLTELDGIEPLRDVVVLGATNRPDLID
Mvan_0749|M.vanbaalenii_PYR-1 IDALAPRRGQSFDSGVTDRVVAALLTELDGIEPLVDVVVLGATNRPDLID
TH_2865|M.thermoresistible__bu IDALAPRRGQSVDSGVTDRVVASLLTELDGIEPLRDVVVLGATNRPDLID
MAB_0637c|M.abscessus_ATCC_199 VDALAPRRGQSFDSGVTDRVVAALLTELDGVEPLRDVVVLGATNRPELID
MSMEG_0858|M.smegmatis_MC2_155 IDALAPRRGQNFDSGVTDKVVASLLTELDGIEPLRDVVVLGATNRPDLID
MLBr_01316|M.leprae_Br4923 LTEFDGDRAACIKA-MIEKVVDRMYAEID--DNRFLEVTYANGDKEVMYF
: : *. ..: : ::** : :*:* : * . :: :
Mb0443c|M.bovis_AF2122/97 PALLRPGRLERLVFVEPPDAAARREILRTAGKSIPLSSD----VDLDEVA
Rv0435c|M.tuberculosis_H37Rv PALLRPGRLERLVFVEPPDAAARREILRTAGKSIPLSSD----VDLDEVA
MMAR_0752|M.marinum_M PALLRPGRLERLVFVEPPDAEARLEILRTAGKSIPLSAD----VDLDEVA
MUL_1384|M.ulcerans_Agy99 PALLRPGRLERLVFVEPPDAEARLEILRTAGKSIPLSAD----VDLDEVA
MAV_4716|M.avium_104 PALLRPGRLERLVFVEPPDAAARREILRTAGKSIPLSAD----VDLDELA
Mflv_0155|M.gilvum_PYR-GCK PALLRPGRLEKLVFVEPPDAEARTQILRTAGKSVPLAAD----VDLEVLA
Mvan_0749|M.vanbaalenii_PYR-1 PALLRPGRLEKLVFVEPPDAEARTQILKTAGKSVPLASDGPEAVDLETLS
TH_2865|M.thermoresistible__bu PALLRPGRLERLVFVEPPDAAARREILATAGKSIPLATEGPDAVDLDALA
MAB_0637c|M.abscessus_ATCC_199 PALLRPGRMERLVFVEPPDAQARTDILRTAARSVPLGAD----VDLNALA
MSMEG_0858|M.smegmatis_MC2_155 PALLRPGRLERLVFVEPPDAAARRDILRTAGKSIPLADD----VDLDSLA
MLBr_01316|M.leprae_Br4923 KDFNSGAMIQNVVDRAKKNAIKSVLETGQPGLRIQHLLD----SIVDEFA
: . ::.:* :* .. : : :: .:
Mb0443c|M.bovis_AF2122/97 AGLDGYSAADCVALLREAALTAMRRSIDAANVTAADLATAR-ETVRASLD
Rv0435c|M.tuberculosis_H37Rv AGLDGYSAADCVALLREAALTAMRRSIDAANVTAADLATAR-ETVRASLD
MMAR_0752|M.marinum_M AGLDGYSAADCVALLREAALTAMRRSIDAADVTAADLAAAR-QTVRPSLD
MUL_1384|M.ulcerans_Agy99 AGLDGYSAADCVALLREAALTAMRRSIDAADVTAADLAAARRQTVRPSLD
MAV_4716|M.avium_104 AGLDGYSAADCVALLREAALTAMRRSIDATDVTAADLAAAR-ENVRPSLD
Mflv_0155|M.gilvum_PYR-GCK ADLDGYSAADCVALLREAALTAMRRSIDAADVTADDVARAR-ETVRPSLD
Mvan_0749|M.vanbaalenii_PYR-1 ADLDGYSAADCVALLREAALTAMRRSIDAADVTASDVATAR-QTVRPSLD
TH_2865|M.thermoresistible__bu AELDGYSAADCVALLREAALTAMRRSIDAADVTAADVARAR-ETVRPSLD
MAB_0637c|M.abscessus_ATCC_199 ADLDGFSAADCAALLREAAMAAMRRSIDAADVTSADIHTAR-SVVRPSLD
MSMEG_0858|M.smegmatis_MC2_155 DDLDGYSAADCVALLRESAMTAMRRSIDAADVTAADVAKAR-ETVRPSLD
MLBr_01316|M.leprae_Br4923 ENEDLPNTTNPDDWARISGKKGERIVYIRTLVTGKSSSASRAIDTESNLG
* .::: * :. . * : **. . :* ....*.
Mb0443c|M.bovis_AF2122/97 PLQVASLRKFGTKGDLRS
Rv0435c|M.tuberculosis_H37Rv PLQVASLRKFGTKGDLRS
MMAR_0752|M.marinum_M PLQVDALRAFGEEL----
MUL_1384|M.ulcerans_Agy99 PLQVDALRAFGEEL----
MAV_4716|M.avium_104 PAQVASLRAFAEAP----
Mflv_0155|M.gilvum_PYR-GCK PAQVESLRQFADTR----
Mvan_0749|M.vanbaalenii_PYR-1 PVQVESLRAFAESR----
TH_2865|M.thermoresistible__bu PAQVQALRAFAAGR----
MAB_0637c|M.abscessus_ATCC_199 PVQVASLRAFADNR----
MSMEG_0858|M.smegmatis_MC2_155 PAQVESLREFAEKR----
MLBr_01316|M.leprae_Br4923 QYL---------------