For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VTPVPDFIVELRRRIGHAPLWLPGITAVTIRGRKVLLVKRSDNGAWTAVTGIVEPGENPADCAAREVREE TGVSARATRLAWVHVTRPAIHANGDHAQYLDHVFRMEWLSGEPFPADDESTAAAWFDLDKLPPMTADMRR RITLSANDDERTVFDTDGPPPARPSG
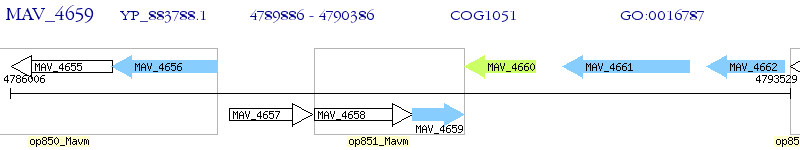
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_4659 | - | - | 100% (166) | hydrolase, nudix family protein, putative |
| M. avium 104 | MAV_3798 | - | 7e-10 | 34.92% (126) | nudix hydrolase |
| M. avium 104 | MAV_5296 | - | 3e-06 | 29.01% (131) | MutT/nudix family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3938 | - | 4e-07 | 30.53% (131) | hypothetical protein Mb3938 |
| M. gilvum PYR-GCK | Mflv_3031 | - | 2e-10 | 36.97% (119) | NUDIX hydrolase |
| M. tuberculosis H37Rv | Rv3908 | - | 4e-07 | 30.53% (131) | hypothetical protein Rv3908 |
| M. leprae Br4923 | MLBr_02698 | - | 2e-05 | 28.24% (131) | hypothetical protein MLBr_02698 |
| M. abscessus ATCC 19977 | MAB_1514 | - | 9e-66 | 70.25% (158) | putative MutT/NUDIX-family protein |
| M. marinum M | MMAR_3129 | - | 5e-11 | 37.82% (119) | hypothetical protein MMAR_3129 |
| M. smegmatis MC2 155 | MSMEG_0197 | - | 4e-08 | 33.10% (142) | nudix hydrolase |
| M. thermoresistible (build 8) | TH_0513 | - | 9e-06 | 29.41% (119) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_5061 | - | 3e-08 | 31.11% (135) | hypothetical protein MUL_5061 |
| M. vanbaalenii PYR-1 | Mvan_6062 | - | 5e-06 | 28.68% (136) | NUDIX hydrolase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3031|M.gilvum_PYR-GCK --------------------------------------------------
MMAR_3129|M.marinum_M --------------------------------------------------
MAV_4659|M.avium_104 --------------------------------------------------
MAB_1514|M.abscessus_ATCC_1997 --------------------------------------------------
Mb3938|M.bovis_AF2122/97 -------MSDGEQAKSRRRRGRRRGRRAAATAENHMDAQPAGDATPTPAT
Rv3908|M.tuberculosis_H37Rv -------MSDGEQAKSRRRRGRRRGRRAAATAENHMDAQPAGDATPTPAT
MUL_5061|M.ulcerans_Agy99 -------MSEGEQAKPRRRRGRRRGRGAARSAENHTDN--AGSPDPNPAP
MLBr_02698|M.leprae_Br4923 MPAIIAWVSDGEQAKPRRRRKRRRGRDATRSAENHTDRQVANDATAT---
TH_0513|M.thermoresistible__bu --------------------------------------------------
Mvan_6062|M.vanbaalenii_PYR-1 MPATIAWVSDGEQAKPRRRRGRRRGRRAAGPPEALPDQAADHRQQAPPAG
MSMEG_0197|M.smegmatis_MC2_155 --------MPSDHDASDLIRRQALTLLAVAQNGLTFAHDPFDRQRYTQVR
Mflv_3031|M.gilvum_PYR-GCK -------------------MRIDYYNDPNAPAPNSVVPSASAIVTDEHGR
MMAR_3129|M.marinum_M -------------------MRTDYYNDPNAPLPNSVVPSASAIVTDEQGR
MAV_4659|M.avium_104 ------MTPVPD--------FIVELRRRIGHAPLWLPGITAVTIRGRKVL
MAB_1514|M.abscessus_ATCC_1997 -------MPVPE--------FVLELRRHIGHAPLWLPGVTGVVIRGEQVL
Mb3938|M.bovis_AF2122/97 AKRSRSRSPRRG-------STRMRTVHETSAGGLVIDGIDGPRDAQVAAL
Rv3908|M.tuberculosis_H37Rv AKRSRSRSPRRG-------STRMRTVHETSAGGLVIDGIDGPRDAQVAAL
MUL_5061|M.ulcerans_Agy99 TKGRRPRLPRRGR------PDRLRTVHETSAGGLVIDGIDGPRDAQVAAL
MLBr_02698|M.leprae_Br4923 -KPRRSHSPHRA-------PEQLRTVHETSAGGLVIYGIDGPRDAQVAAL
TH_0513|M.thermoresistible__bu ----------------------LRTVHETSAGGLVIDGLDGPKEKQVAAL
Mvan_6062|M.vanbaalenii_PYR-1 TAGQARQTPKPQRTRTRRPPERLRTVHETSAGGLVIDGIDGPKDGQVAAL
MSMEG_0197|M.smegmatis_MC2_155 RAAEELMTLIADG-----DIEQLRAAFSVDTGYMTPKVSVRGAIFNSEEE
Mflv_3031|M.gilvum_PYR-GCK ILLIKRRDNTLWALPGGGHDIGETIADTAVREVKEETGLDVEVTGLVGVY
MMAR_3129|M.marinum_M ILLIKRRDNTLWALPGGGHDIGETIADTAVREVKEETGLDIEVTGLVGVY
MAV_4659|M.avium_104 LVKRSDNG--AWTAVTGIVEPGENPADCAAREVREETGVSARATRLAWVH
MAB_1514|M.abscessus_ATCC_1997 LVKRADNG--AWTAVTGIVDPGENPADCASREVLEETGVRATPRRLVWVH
Mb3938|M.bovis_AF2122/97 IGRVDRRGRLLWSLPKGHIELGETAEQTAIREVAEETGIRG--SVLAALG
Rv3908|M.tuberculosis_H37Rv IGRVDRRGRLLWSLPKGHIELGETAEQTAIREVAEETGIRG--SVLAALG
MUL_5061|M.ulcerans_Agy99 IGRIDRRGRMLWSLPKGHIEMGETAEQTAIREVAEETGIRG--GVLAALG
MLBr_02698|M.leprae_Br4923 IGRIDRRGRMLWSLPKGHIEQGETAEQTAIREVAEETGIRG--SVLAALG
TH_0513|M.thermoresistible__bu IGRLDRRGRMLWSLPKGHIEMGETAEQTAIREVAEETGVQG--AVLAALG
Mvan_6062|M.vanbaalenii_PYR-1 IGRIDRRGRMLWSLPKGHIEMGETAEQTAIREVAEETGIQG--SVLAALG
MSMEG_0197|M.smegmatis_MC2_155 LLLVQERADRLWTLPGGWCDVLETPAQAVAKEVREEAGLIVDVDKLVAVL
: . *: * : *. : . :** **:*: *. :
Mflv_3031|M.gilvum_PYR-GCK TNPHHVVAFTDGEVRQQFSLSFTTKVLGG-TLAIDHESTDIAWTSPDDIP
MMAR_3129|M.marinum_M TNPQHVVAFTDGEVRQQFSLSFTTKVLGG-TLAIDHESTDIAWTDPDDIP
MAV_4659|M.avium_104 V--TRPAIHANGDHAQYLDHVFRMEWLSGEPFPADDESTAAAWFDLDKLP
MAB_1514|M.abscessus_ATCC_1997 V--SRPIVHVNGDHAQYLDHVFRMDWVAGSPFPADDENDAAQWFDITAMP
Mb3938|M.bovis_AF2122/97 R--IDYWFVTDGRRVHKTVHHYLMRFLGGELSDEDLEVAEVAWVPIRELP
Rv3908|M.tuberculosis_H37Rv R--IDYWFVTDGRRVHKTVHHYLMRFLGGELSDEDLEVAEVAWVPIRELP
MUL_5061|M.ulcerans_Agy99 R--IDYWFVTDGRRVHKTVHHYLMRFLGGELSDNDLEVTEVAWVPIPELP
MLBr_02698|M.leprae_Br4923 Q--IDYWFVTDDCRVHKTVHHYLMRFSGGELSDDDLEVTEVAWVPIRELP
TH_0513|M.thermoresistible__bu S--IDYWFVTEGRRVHKTVHHYLMRFLGGELSDDDVEVSEVAWVPLSELP
Mvan_6062|M.vanbaalenii_PYR-1 S--IDYWFVTEGRRVHKTVHHYLMRFSGGELSDEDVEVTEVAWVPVKDLP
MSMEG_0197|M.smegmatis_MC2_155 YH-DRHRPSRQPAPLFHVHKLFFLCHERGRVPADLTGTSAIDWFALDRLP
: : * * :*
Mflv_3031|M.gilvum_PYR-GCK SLDMHPSMRLRIEHYLQHRDNPYLG-------------------------
MMAR_3129|M.marinum_M NLDMHPSMRLRIEHYLQHRDSPYLG-------------------------
MAV_4659|M.avium_104 P--MTADMRRRITLSAND-DERTVFDTDGPPPARPSG-------------
MAB_1514|M.abscessus_ATCC_1997 E--MTADMRRRIELALDIGSAETVFEVTEP-----SG-------------
Mb3938|M.bovis_AF2122/97 SRLAYADERRLAEVADELIDKLQSDGPAALPPLPPSSPRRRPQTHSRAR-
Rv3908|M.tuberculosis_H37Rv SRLAYADERRLAEVADELIDKLQSDGPAALPPLPPSSPRRRPQTHSRAR-
MUL_5061|M.ulcerans_Agy99 SRLAYADERRLAEVADELIDKLQTDGPTALPPLPPSAPRRRPQTHSRTR-
MLBr_02698|M.leprae_Br4923 SRLAYADERRLAEVAYELINKLQTGGPAALPPLPPSLPRRQSQTHFRTR-
TH_0513|M.thermoresistible__bu TRLAYADERRLAEVAGELIDTLQTHGPQALPPLPQMAPRRRPQTHSRTRS
Mvan_6062|M.vanbaalenii_PYR-1 SRLAYADERRLAEVADELIDKLHTDGPGALPPLPRTTPRRRGQTHSHTR-
MSMEG_0197|M.smegmatis_MC2_155 PLAPSVDEAQLRMMHTHWRHPELPTVFD----------------------
. .
Mflv_3031|M.gilvum_PYR-GCK -----------------------------------
MMAR_3129|M.marinum_M -----------------------------------
MAV_4659|M.avium_104 -----------------------------------
MAB_1514|M.abscessus_ATCC_1997 -----------------------------------
Mb3938|M.bovis_AF2122/97 --HADD-SAPGQ---------------HNGPGPGP
Rv3908|M.tuberculosis_H37Rv --HADD-SAPGQ---------------HNGPGPGP
MUL_5061|M.ulcerans_Agy99 --HAAEGSPPGR---------------KNGHGPGP
MLBr_02698|M.leprae_Br4923 --HSNN-SANSR---------------KNGHGQGP
TH_0513|M.thermoresistible__bu SRRRDDSAAHSRGEGRERGAANSSGRRTNGCGKGP
Mvan_6062|M.vanbaalenii_PYR-1 -RHRPDPTAQPQ-----------PGRRTNGCGQGP
MSMEG_0197|M.smegmatis_MC2_155 -----------------------------------