For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
PSDHDASDLIRRQALTLLAVAQNGLTFAHDPFDRQRYTQVRRAAEELMTLIADGDIEQLRAAFSVDTGYM TPKVSVRGAIFNSEEELLLVQERADRLWTLPGGWCDVLETPAQAVAKEVREEAGLIVDVDKLVAVLYHDR HRPSRQPAPLFHVHKLFFLCHERGRVPADLTGTSAIDWFALDRLPPLAPSVDEAQLRMMHTHWRHPELPT VFD*
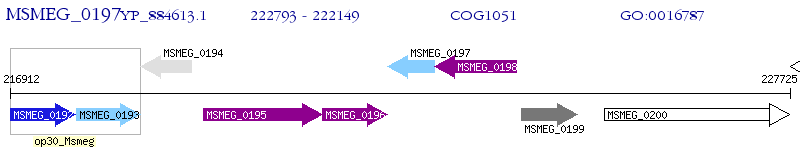
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_0197 | - | - | 100% (214) | nudix hydrolase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_3031 | - | 1e-08 | 30.66% (137) | NUDIX hydrolase |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_2096c | - | 3e-08 | 38.81% (67) | putative MutT/NUDIX-like protein |
| M. marinum M | MMAR_3129 | - | 2e-08 | 29.93% (137) | hypothetical protein MMAR_3129 |
| M. avium 104 | MAV_3798 | - | 2e-08 | 38.81% (67) | nudix hydrolase |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_0524 | - | 9e-05 | 32.88% (73) | hypothetical protein MUL_0524 |
| M. vanbaalenii PYR-1 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3031|M.gilvum_PYR-GCK --------------------------------------------------
MMAR_3129|M.marinum_M --------------------------------------------------
MAB_2096c|M.abscessus_ATCC_199 --------------------------------------------------
MAV_3798|M.avium_104 --------------------------------------------------
MUL_0524|M.ulcerans_Agy99 --------------------------------------------------
MSMEG_0197|M.smegmatis_MC2_155 MPSDHDASDLIRRQALTLLAVAQNGLTFAHDPFDRQRYTQVRRAAEELMT
Mflv_3031|M.gilvum_PYR-GCK ---------MRIDYYNDPNAPAPNSVVPSASAIVTDEHGRILLIKRRDNT
MMAR_3129|M.marinum_M ---------MRTDYYNDPNAPLPNSVVPSASAIVTDEQGRILLIKRRDNT
MAB_2096c|M.abscessus_ATCC_199 ---------MRTDYYNDPNAPAPNSVVPSASAIVTDEQGRILLIKRRDNT
MAV_3798|M.avium_104 ---------MRTDYYNDPNAPRPNSVVPSASAIVADERGRILLIKRRDNT
MUL_0524|M.ulcerans_Agy99 ---------MR-------MAATPKHSVSVAGIVVRDD-GRVLVIKRDDNG
MSMEG_0197|M.smegmatis_MC2_155 LIADGDIEQLRAAFSVDTGYMTPK--VSVRGAIFNSE-EELLLVQERADR
:* *: *. . :. .: .:*:::. :
Mflv_3031|M.gilvum_PYR-GCK LWALPGGGHDIGETIADTAVREVKEETGLDVEVTGLVGVYTNPHHVVAFT
MMAR_3129|M.marinum_M LWALPGGGHDIGETIADTAVREVKEETGLDIEVTGLVGVYTNPQHVVAFT
MAB_2096c|M.abscessus_ATCC_199 LWALPGGGHDIGETIADTAVREVKEETGLDVEVTGLVGVYTNPHHVVAFT
MAV_3798|M.avium_104 LWALPGGGHDIGETIEQTAVREVKEETGLDVEITGLVGVYTNPRHVVAFT
MUL_0524|M.ulcerans_Agy99 HWEAPGGVLELDESFEAGVQREVLEETGVEVTVERLTGVYKN--------
MSMEG_0197|M.smegmatis_MC2_155 LWTLPGGWCDVLETPAQAVAKEVREEAGLIVDVDKLVAVLYHDRHRPSRQ
* *** :: *: . :** **:*: : : *..* :
Mflv_3031|M.gilvum_PYR-GCK DGEVRQQFSLSFTTKVLGGTLAIDHESTDIAWTSPDDIPSLDMHPSMRLR
MMAR_3129|M.marinum_M DGEVRQQFSLSFTTKVLGGTLAIDHESTDIAWTDPDDIPNLDMHPSMRLR
MAB_2096c|M.abscessus_ATCC_199 DGEVRQQFSLLFATTVLGGTLAIDHESTDIAWTAPDDIAGLDMHPSMRLR
MAV_3798|M.avium_104 DGEVRQQFSLLFTTRVLGGELAIDHESTDIAWTDPDDIADLDMHPSMRLR
MUL_0524|M.ulcerans_Agy99 --LAHGIVALVYRCRPLGDEAHATEEACEIRWMTKEEVQS-AMVPAFGVR
MSMEG_0197|M.smegmatis_MC2_155 PAPLFHVHKLFFLCHERGRVPADLTGTSAIDWFALDRLPPLAPSVDEAQL
* : * : * * : :
Mflv_3031|M.gilvum_PYR-GCK --IEHYLQHRDNPYLG--------
MMAR_3129|M.marinum_M --IEHYLQHRDSPYLG--------
MAB_2096c|M.abscessus_ATCC_199 --VEHYLQRRDSPYLG--------
MAV_3798|M.avium_104 --IEHYLQHRDSPYLC--------
MUL_0524|M.ulcerans_Agy99 --VLDAFEEAPQSRAHDGVNLVRV
MSMEG_0197|M.smegmatis_MC2_155 RMMHTHWRHPELPTVFD-------
: .. .