For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTRTHDDEWDLASSVGATATMVAAGRAMATKDPRGLIDDPFAEPLVRAVGVDFFTKMMDGELDLDAIENA TPVRIQSMVDGMAVRTKYFDDYFVDATDAGVRQVVILASGLDSRAYRLPWPAGTVVYEIDQPRVIEFKSN TLAEVGAEPTATRRTIPIDLRGDWPAALSAAGFDPAAPTAWLAEGLLIYLPPEAQDRLFDNITALSAPGS TIATEFVPGIVDFDAERVREMSGSFREHGVDIDMASLVYAGERNHVIDYLNGLGWRAEGVTRTELFHRHG IEVPAPEHDDPLGEIIFISATRTG
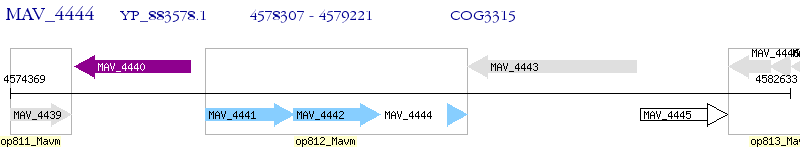
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_4444 | - | - | 100% (304) | methyltransferase, putative, family protein |
| M. avium 104 | MAV_4441 | - | 5e-95 | 57.52% (306) | methyltransferase, putative, family protein |
| M. avium 104 | MAV_4442 | - | 1e-94 | 58.22% (304) | methyltransferase, putative, family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1758c | - | 1e-120 | 69.64% (303) | hypothetical protein Mb1758c |
| M. gilvum PYR-GCK | Mflv_5025 | - | 1e-118 | 67.55% (302) | putative methyltransferase |
| M. tuberculosis H37Rv | Rv1729c | - | 1e-120 | 69.64% (303) | hypothetical protein Rv1729c |
| M. leprae Br4923 | MLBr_02640 | - | 3e-68 | 49.36% (312) | hypothetical protein MLBr_02640 |
| M. abscessus ATCC 19977 | MAB_3787 | - | 1e-90 | 56.11% (303) | hypothetical protein MAB_3787 |
| M. marinum M | MMAR_1057 | - | 1e-143 | 79.28% (304) | O-methyltransferase |
| M. smegmatis MC2 155 | MSMEG_1479 | - | 1e-113 | 64.78% (301) | methyltransferase, putative, family protein |
| M. thermoresistible (build 8) | TH_2706 | - | 2e-94 | 59.51% (284) | PUTATIVE putative methyltransferase |
| M. ulcerans Agy99 | MUL_0816 | - | 1e-142 | 78.62% (304) | O-methyltransferase |
| M. vanbaalenii PYR-1 | Mvan_1344 | - | 1e-120 | 69.97% (303) | putative methyltransferase |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_1057|M.marinum_M MARTHDDKWDLASSVGATATMVAAGRAMASRDPRGLIDDPFAEPLVRAVG
MUL_0816|M.ulcerans_Agy99 MARTHDDKWDLASSVGATATIVAAGRAMASRDPRGLIDDPFAEPLVRAVG
MAV_4444|M.avium_104 MTRTHDDEWDLASSVGATATMVAAGRAMATKDPRGLIDDPFAEPLVRAVG
Mb1758c|M.bovis_AF2122/97 MARTDDDNWDLTSSVGVTATIVAVGRALATKDPRGLINDPFAEPLVRAVG
Rv1729c|M.tuberculosis_H37Rv MARTDDDNWDLTSSVGVTATIVAVGRALATKDPRGLINDPFAEPLVRAVG
Mflv_5025|M.gilvum_PYR-GCK MARTPDDSWDLASSVGATATMVAAGRAVASADPDPLINDPYAEPLVRAVG
Mvan_1344|M.vanbaalenii_PYR-1 MARTADDSWDPASSVGATATMVAAGRAAASADPDPLINDPYAEPLVRAVG
MSMEG_1479|M.smegmatis_MC2_155 MVRSDNDTWDLASSVGATATMVAAGRAVVSQDPGGLINDPFAAPLVRAVG
MAB_3787|M.abscessus_ATCC_1997 MARTDDDSWDLASSVGATATMVAAQRALASHVPNPLINDPYAEPLVRAVG
TH_2706|M.thermoresistible__bu MARSESDTWDLTSGVGVTATMVAAARALASREPEPLIDDPFAAPLVRAVG
MLBr_02640|M.leprae_Br4923 -MRTHDDTWDIKTSVGTTAVMVAAARAAETDRPDALIRDPYAKLLVTNTG
*: .* ** :.**.**.:**. ** : * ** **:* ** .*
MMAR_1057|M.marinum_M V-DFFIKMMDGEFDLSVLQNVSSA--KAQAMVDGMAVRTKYFDDYFGDAI
MUL_0816|M.ulcerans_Agy99 V-DFFIKMMDGEFDLSVLQNVSSA--KAQAMVDGMAVRTKYFDDYFGDAI
MAV_4444|M.avium_104 V-DFFTKMMDGELDLDAIENATPV--RIQSMVDGMAVRTKYFDDYFVDAT
Mb1758c|M.bovis_AF2122/97 L-DLFTKMMDGELDMSTIADVSPA--VAQAMVYGNAVRTKYFDDYLLNAT
Rv1729c|M.tuberculosis_H37Rv L-DLFTKMMDGELDMSTIADVSPA--VAQAMVYGNAVRTKYFDDYLLNAT
Mflv_5025|M.gilvum_PYR-GCK L-DFFTRMLDGELDLSVFPDSSPE--RAQAMIDGMAVRTRFFDDCCLAAA
Mvan_1344|M.vanbaalenii_PYR-1 L-SFFTKMLDGELDLSQFADGSPE--RVQAMIDGMAVRTRFFDDCCGTST
MSMEG_1479|M.smegmatis_MC2_155 I-EALTMLADGKFDIEKVLPESAA--RVRANIDEMAVRTKFFDDYFMDAT
MAB_3787|M.abscessus_ATCC_1997 I-EYFTKLASGAFDPEQMAGLVQLGITADGMANGMASRTWYYDTAFESST
TH_2706|M.thermoresistible__bu V-EFFTRLVDGDLELPADTAEERA--AAQVLTEVMAVRTRFFDDFFTDAT
MLBr_02640|M.leprae_Br4923 AGALWEAMLDPSMVAKVEAIDAEAAAMVEHMRSYQAVRTNFFDTYFNNAV
: . : * ** ::* :
MMAR_1057|M.marinum_M KSGIRQAVILASGLDARAYRLPWPADTVVYELDQPQVIEFKTNVLADLGA
MUL_0816|M.ulcerans_Agy99 KSGIRQAVILASGLDARAYRLPWPADTVVYELDQPQVIEFKTNVLADLGA
MAV_4444|M.avium_104 DAGVRQVVILASGLDSRAYRLPWPAGTVVYEIDQPRVIEFKSNTLAEVGA
Mb1758c|M.bovis_AF2122/97 AGGIRQVAILASGLDSRAYRLPWPTRTVVYEIDQPKVMEFKTTTLADLGA
Rv1729c|M.tuberculosis_H37Rv AGGIRQVAILASGLDSRAYRLPWPTRTVVYEIDQPKVMEFKTTTLADLGA
Mflv_5025|M.gilvum_PYR-GCK SAGVRQVVILAAGLDARTYRLPWPDGTVVYELDQPDVIAFKTQTLRNLGA
Mvan_1344|M.vanbaalenii_PYR-1 TAGITQVVILASGLDARAYRLGWPAGTVVYELDQPAVIEFKTRTLAGLGA
MSMEG_1479|M.smegmatis_MC2_155 GRGVGQAVILASGLDSRAYRLPWPDGTVVYEIDQPDVIEFKTRTLADLGA
MAB_3787|M.abscessus_ATCC_1997 AAGVRQAVILASGLDTRAYRLTWPAGTTVYELDQPEVITFKTDTLAELGA
TH_2706|M.thermoresistible__bu RVGARQAVILASGLDARAYRLTWPDGTVVYEIDQPEVIAFKTRTLAELGA
MLBr_02640|M.leprae_Br4923 IDGIRQFVILASGLDSRAYRLDWPTGTTVYEIDQPKVLAYKSTTLAEHGV
* * .***:***:*:*** ** *.***:*** *: :*: .* *.
MMAR_1057|M.marinum_M EPRATRRAIPIDLRGDWPVALRAAGLDTTAPTAWLAEGLLIYLPPEAQDR
MUL_0816|M.ulcerans_Agy99 EPRATRRAIPIDLRGDWPVALRAAGLDTTAPTAWLAEGLLIYLPPEAQDR
MAV_4444|M.avium_104 EPTATRRTIPIDLRGDWPAALSAAGFDPAAPTAWLAEGLLIYLPPEAQDR
Mb1758c|M.bovis_AF2122/97 EPSAIRRAVPIDLRADWPTALQAAGFDSAAPTAWLAEGLLIYLKPQTQDR
Rv1729c|M.tuberculosis_H37Rv EPSAIRRAVPIDLRADWPTALQAAGFDSAAPTAWLAEGLLIYLKPQTQDR
Mflv_5025|M.gilvum_PYR-GCK EPAATQRPVPVDLREDWPAALRAAGFDATRPTAWLAEGLLIYLPPEAQDA
Mvan_1344|M.vanbaalenii_PYR-1 HPTATHRPVPIDLREDWPAALHAAGFDASRPTAWLAEGLLIYLPPEAQDQ
MSMEG_1479|M.smegmatis_MC2_155 EPTCERRTVSIDLRDDWPAALRAAGFDPSAPTAWCAEGLLIYLPPEAQDK
MAB_3787|M.abscessus_ATCC_1997 SPSAERRTVAIDLREDWPAALQAAGFDPEQPTVWSAEGLLIYLPPEAQDR
TH_2706|M.thermoresistible__bu PPTAPHRPVAVDLRDDWPAALTAAGFDPTVATAWSAEGLLRYLPPDAQDR
MLBr_02640|M.leprae_Br4923 TPTADRREVPIDLRQDWPPALRSAGFDPSARTAWLAEGLLMYLPATAQDG
* . :* :.:*** *** ** :**:*. *.* ***** ** . :**
MMAR_1057|M.marinum_M LFDNITALSAPGSTVATEFVPGIVDFDVDRARQMSGPFRDHG---LDIDM
MUL_0816|M.ulcerans_Agy99 LFDNITALSAPGSTVATEFVPGIVDFDVDRARQMSGPFRDHG---LDIDM
MAV_4444|M.avium_104 LFDNITALSAPGSTIATEFVPGIVDFDAERVREMSGSFREHG---VDIDM
Mb1758c|M.bovis_AF2122/97 LFDNITALSAPGSMVATEFVTGIADFSAERARTISNPFRCHG---VDVDL
Rv1729c|M.tuberculosis_H37Rv LFDNITALSAPGSMVATEFVTGIADFSAERARTISNPFRCHG---VDVDL
Mflv_5025|M.gilvum_PYR-GCK LFDTVTALSAPGSTLATEYVPGIVDFDGAKARALSEPLREHG---LDIDM
Mvan_1344|M.vanbaalenii_PYR-1 LFDRITALSAPGSTIATEYAPGIIDFDDEKARAMSAPMREMG---LDIDM
MSMEG_1479|M.smegmatis_MC2_155 LFDDIHHLSAPGSTVATEFVPALKDFDPEKARQATETLTRMG---VDIDM
MAB_3787|M.abscessus_ATCC_1997 LFASITELSAPGSRLVCEQVPGLETADFSKARELTRQFAGET---LDLDL
TH_2706|M.thermoresistible__bu LFDHITDLSAPGSRLATEDHPGPGAGLGEKADELGRRWREHG---FEVQL
MLBr_02640|M.leprae_Br4923 LFTEIGGLSAVGSRIAVETSPLHGDEWREQMQLRFRRVSDALGFEQAVDV
** : *** ** :. * . : :::
MMAR_1057|M.marinum_M SSLVYTGA-RNHVVDYLRAKGWDAEGVTRSELFERNGMA---VPAPSDD-
MUL_0816|M.ulcerans_Agy99 SSLVYTGA-RNHVVDYLRAKGWDAEGVTRSKLFERNGMA---VPAPSDD-
MAV_4444|M.avium_104 ASLVYAGE-RNHVIDYLNGLGWRAEGVTRTELFHRHGIE---VPAPEHD-
Mb1758c|M.bovis_AF2122/97 ASLVYTGP-RNHVLDYLAAKGWQPEGVSLAELFRRSGLD---VRAADDDT
Rv1729c|M.tuberculosis_H37Rv ASLVYTGP-RNHVLDYLAAKGWQPEGVSLAELFRRSGLD---VRAADDDT
Mflv_5025|M.gilvum_PYR-GCK PSLVYTGA-RTPVMDHLRGAGWQVSGTARAELFARFGRP---LPQDADG-
Mvan_1344|M.vanbaalenii_PYR-1 PSLVYAGT-RSHVMDYLAGQGWEVTGVPRRELFTRYGRP---LPPEAGD-
MSMEG_1479|M.smegmatis_MC2_155 PSLIYHGE-RHSASDYLETKGWQMDSAARATLFTRYGLP---APGHDDD-
MAB_3787|M.abscessus_ATCC_1997 ESLVYTQE-RQMAADWLAEHGWTVITEENDELYARLNLD---PPNPLLR-
TH_2706|M.thermoresistible__bu SELFHPGE-RTPVVDYLRGRGWDARARPRAEVFAGYGRE---FPDSAAL-
MLBr_02640|M.leprae_Br4923 QELIYHDENRAVVADWLNRHGWRATAQSAPDEMRRVGRWGDGVPMADDK-
.*.: * . * * ** .
MMAR_1057|M.marinum_M ----DPLGEIIFISAALTG------
MUL_0816|M.ulcerans_Agy99 ----DPLGEIIFISAALTG------
MAV_4444|M.avium_104 ----DPLGEIIFISATRTG------
Mb1758c|M.bovis_AF2122/97 IFISGCLTDHSSISPPTAAGWR---
Rv1729c|M.tuberculosis_H37Rv IFISGCLTDHSSISPPTAAGWR---
Mflv_5025|M.gilvum_PYR-GCK ---TDPLGEIVYVSATLAADA----
Mvan_1344|M.vanbaalenii_PYR-1 ---TDPLGEIVYVSAHRPQ------
MSMEG_1479|M.smegmatis_MC2_155 ----DPLGEIIYISGTLR-------
MAB_3787|M.abscessus_ATCC_1997 ----SIFPNIVYVDATLG-------
TH_2706|M.thermoresistible__bu ----AAMRDSSAVTAVRPVVRAGMR
MLBr_02640|M.leprae_Br4923 ----DAFAEFVTAHRL---------
: :