For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTKLAIIYYSATGHGTTMARRVAAAAESAGAQVRLRHVAETQDPESFAHNPAWTANYQATKDLPAATGDD IVWADAVIFGSPTRFGSPAAQLRAFLDSLGGLWAQGKLADKVYAAFTSSNTLHGGQETTLLSLYITLMHF GGIIVAPGYTDPSKFVDGNPYGASLVTTHDNIHEFDEPTANALDHLARRVVETAGRLAS
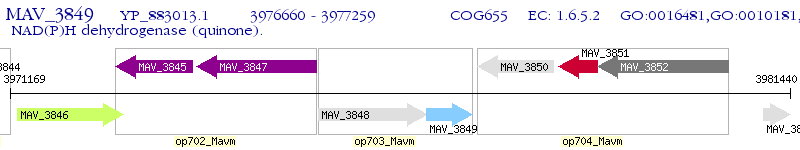
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_3849 | wrbA | - | 100% (199) | NAD(P)H:quinone oxidoreductase, type IV |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_3384 | - | 2e-05 | 28.65% (171) | hypothetical protein Mflv_3384 |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | MMAR_1713 | - | 1e-101 | 87.44% (199) | hypothetical protein MMAR_1713 |
| M. smegmatis MC2 155 | MSMEG_2375 | wrbA | 2e-92 | 80.20% (197) | NAD(P)H:quinone oxidoreductase, type IV |
| M. thermoresistible (build 8) | TH_0039 | - | 1e-11 | 28.99% (207) | flavodoxin/nitric oxide synthase |
| M. ulcerans Agy99 | MUL_1950 | - | 1e-100 | 86.43% (199) | hypothetical protein MUL_1950 |
| M. vanbaalenii PYR-1 | Mvan_2126 | - | 4e-88 | 77.66% (197) | flavodoxin/nitric oxide synthase |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_1713|M.marinum_M --------MTKLAVIYYSATGHGTTMARHVAAAAESAGAEVRVRHVAETR
MUL_1950|M.ulcerans_Agy99 --------MTKLAVIYYSATGHGTTMARHVAAAAESAGAEVRVRHVAETR
MAV_3849|M.avium_104 --------MTKLAIIYYSATGHGTTMARRVAAAAESAGAQVRLRHVAETQ
MSMEG_2375|M.smegmatis_MC2_155 ----MTSSATKVAVIYYSATGHGTTMAQRVAASAEAAGAEVRLRHIAETQ
Mvan_2126|M.vanbaalenii_PYR-1 --------MTKVAVIYYSATGHGTAMANRVAAAAEAAGAEVRVRHVAETR
TH_0039|M.thermoresistible__bu -----------LAIAYHSGFGHTARLAEAVAAGARESGADVTVLPVDALT
Mflv_3384|M.gilvum_PYR-GCK MNSAGESNGTRVLLVYYSYTGQTQKVLDAAADVLRERGCEVTLAPIEFVD
: : *:* *: : .* . *.:* : :
MMAR_1713|M.marinum_M -----DPDSFAHNPAWSANYEATKDLPAATGDDIVWADAV--------IF
MUL_1950|M.ulcerans_Agy99 -----DPDSSAHNPAWSANYEATKDLPAATGDDIVWADAV--------IF
MAV_3849|M.avium_104 -----DPESFAHNPAWTANYQATKDLPAATGDDIVWADAV--------IF
MSMEG_2375|M.smegmatis_MC2_155 -----DPQTFTQNPAWTANYEATKDLPAATGEDIVWADAV--------VF
Mvan_2126|M.vanbaalenii_PYR-1 -----DPETFANNPAWTANYEATKDLPAATGEDIVWADAV--------IF
TH_0039|M.thermoresistible__bu -----EQG-------WAV---------------LDGADGI--------IF
Mflv_3384|M.gilvum_PYR-GCK PQYSEPFTRFPMRSVWPDMFSVYKAQGRGETGEIRTPDAVRDGRYDLVIF
*. : .*.: :*
MMAR_1713|M.marinum_M GSPTRFGSVAAQLREFLDSLGGLWAEGKLADKIYAAYTSTNTAHGGQETT
MUL_1950|M.ulcerans_Agy99 GSPTRFGSVAAQLREFLDSLGGLWAEGKLADKIYAAYTSTNTAHGGQETT
MAV_3849|M.avium_104 GSPTRFGSPAAQLRAFLDSLGGLWAQGKLADKVYAAFTSSNTLHGGQETT
MSMEG_2375|M.smegmatis_MC2_155 GSPTRFGSPAAQFRAFIDSLGGLWAQGRLADKVYAAFTSSQTAHGGQEST
Mvan_2126|M.vanbaalenii_PYR-1 GSPTRFGNTASQFRTFVDSLGGLWAQGKLADKVYAGFTSSQTAHGGQETT
TH_0039|M.thermoresistible__bu GTPTYMGNVSAAFQAFTEQTGRRCQEGTWRDKVAAGFTNSGGKSGDKLQT
Mflv_3384|M.gilvum_PYR-GCK GSPTWWDTVSMPLRSFLESGD---AKALLDGTPYAVVTVCRRLWRGNLEA
*:** .. : :: * :. . :. .. * * .: :
MMAR_1713|M.marinum_M LLTLYVT----LMHFGGIIVAPGYTDALKFVD-----GNPYGAS-LVATH
MUL_1950|M.ulcerans_Agy99 LLTLYVT----LMHFGGIIVAPGYTDALKFID-----GNPYGAS-LVATH
MAV_3849|M.avium_104 LLSLYIT----LMHFGGIIVAPGYTDPSKFVD-----GNPYGAS-LVTTH
MSMEG_2375|M.smegmatis_MC2_155 LLNLYVT----LMHFGGIIVPPGYTDEVKFAD-----GNPYGVG-LIANR
Mvan_2126|M.vanbaalenii_PYR-1 LITLYVT----LMHFGGIIVPPGYTDGVKFAD-----GNPYGVG-HITGP
TH_0039|M.thermoresistible__bu LLSLAVFAAQHHMHWVNLGLVAGWNSSAASEQDLNRLGFFIGAGGQTDVD
Mflv_3384|M.gilvum_PYR-GCK VRELADKQGGRFADSIHFEYPGGQLTSLLSLA------SYLGSGEYKQRY
: * . : * * .
MMAR_1713|M.marinum_M DNITD-IDEPTANALDHLARRVVHTADRLASG------------------
MUL_1950|M.ulcerans_Agy99 DNIND-IDEPTANALDHLARRVVHTADRLASG------------------
MAV_3849|M.avium_104 DNIHE-FDEPTANALDHLARRVVETAGRLAS-------------------
MSMEG_2375|M.smegmatis_MC2_155 TNITE-IDEITNNALDHLARRVVTVAGRLAG-------------------
Mvan_2126|M.vanbaalenii_PYR-1 ENRNE-LDEATLAALDHLARRVVGVADRLG--------------------
TH_0039|M.thermoresistible__bu AGPEQ-VHDADIRTCRHLGRRVATVTAQLLAGRAVTAAAADPATASAAAR
Mflv_3384|M.gilvum_PYR-GCK LGVKIPTTNISAGQVDQARSFAGRLADRLFGGR-----------------
. : : . : :*