For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TKLAVIYYSATGHGTTMARHVAAAAESAGAEVRVRHVAETRDPDSFAHNPAWSANYEATKDLPAATGDDI VWADAVIFGSPTRFGSVAAQLREFLDSLGGLWAEGKLADKIYAAYTSTNTAHGGQETTLLTLYVTLMHFG GIIVAPGYTDALKFVDGNPYGASLVATHDNITDIDEPTANALDHLARRVVHTADRLASG*
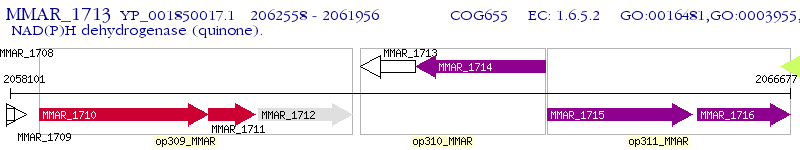
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_1713 | - | - | 100% (200) | hypothetical protein MMAR_1713 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. avium 104 | MAV_3849 | wrbA | 1e-101 | 87.44% (199) | NAD(P)H:quinone oxidoreductase, type IV |
| M. smegmatis MC2 155 | MSMEG_2375 | wrbA | 7e-90 | 78.17% (197) | NAD(P)H:quinone oxidoreductase, type IV |
| M. thermoresistible (build 8) | TH_0039 | - | 4e-12 | 27.62% (210) | flavodoxin/nitric oxide synthase |
| M. ulcerans Agy99 | MUL_1950 | - | 1e-112 | 98.50% (200) | hypothetical protein MUL_1950 |
| M. vanbaalenii PYR-1 | Mvan_2126 | - | 6e-89 | 77.66% (197) | flavodoxin/nitric oxide synthase |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_1713|M.marinum_M ----MTKLAVIYYSATGHGTTMARHVAAAAESAGAEVRVRHVAETRDPDS
MUL_1950|M.ulcerans_Agy99 ----MTKLAVIYYSATGHGTTMARHVAAAAESAGAEVRVRHVAETRDPDS
MAV_3849|M.avium_104 ----MTKLAIIYYSATGHGTTMARRVAAAAESAGAQVRLRHVAETQDPES
MSMEG_2375|M.smegmatis_MC2_155 MTSSATKVAVIYYSATGHGTTMAQRVAASAEAAGAEVRLRHIAETQDPQT
Mvan_2126|M.vanbaalenii_PYR-1 ----MTKVAVIYYSATGHGTAMANRVAAAAEAAGAEVRVRHVAETRDPET
TH_0039|M.thermoresistible__bu -------LAIAYHSGFGHTARLAEAVAAGARESGADVTVLPVDALTEQG-
:*: *:*. ** : :*. ***.*. :**:* : : :
MMAR_1713|M.marinum_M FAHNPAWSANYEATKDLPAATGDDIVWADAVIFGSPTRFGSVAAQLREFL
MUL_1950|M.ulcerans_Agy99 SAHNPAWSANYEATKDLPAATGDDIVWADAVIFGSPTRFGSVAAQLREFL
MAV_3849|M.avium_104 FAHNPAWTANYQATKDLPAATGDDIVWADAVIFGSPTRFGSPAAQLRAFL
MSMEG_2375|M.smegmatis_MC2_155 FTQNPAWTANYEATKDLPAATGEDIVWADAVVFGSPTRFGSPAAQFRAFI
Mvan_2126|M.vanbaalenii_PYR-1 FANNPAWTANYEATKDLPAATGEDIVWADAVIFGSPTRFGNTASQFRTFV
TH_0039|M.thermoresistible__bu ------WAV---------------LDGADGIIFGTPTYMGNVSAAFQAFT
*:. : **.::**:** :*. :: :: *
MMAR_1713|M.marinum_M DSLGGLWAEGKLADKIYAAYTSTNTAHGGQETTLLTLYVT----LMHFGG
MUL_1950|M.ulcerans_Agy99 DSLGGLWAEGKLADKIYAAYTSTNTAHGGQETTLLTLYVT----LMHFGG
MAV_3849|M.avium_104 DSLGGLWAQGKLADKVYAAFTSSNTLHGGQETTLLSLYIT----LMHFGG
MSMEG_2375|M.smegmatis_MC2_155 DSLGGLWAQGRLADKVYAAFTSSQTAHGGQESTLLNLYVT----LMHFGG
Mvan_2126|M.vanbaalenii_PYR-1 DSLGGLWAQGKLADKVYAGFTSSQTAHGGQETTLITLYVT----LMHFGG
TH_0039|M.thermoresistible__bu EQTGRRCQEGTWRDKVAAGFTNSGGKSGDKLQTLLSLAVFAAQHHMHWVN
:. * :* **: *.:*.: *.: **:.* : **: .
MMAR_1713|M.marinum_M IIVAPGYTDALKFVD-----GNPYGAS-LVATHDNITDIDEPTANALDHL
MUL_1950|M.ulcerans_Agy99 IIVAPGYTDALKFID-----GNPYGAS-LVATHDNINDIDEPTANALDHL
MAV_3849|M.avium_104 IIVAPGYTDPSKFVD-----GNPYGAS-LVTTHDNIHEFDEPTANALDHL
MSMEG_2375|M.smegmatis_MC2_155 IIVPPGYTDEVKFAD-----GNPYGVG-LIANRTNITEIDEITNNALDHL
Mvan_2126|M.vanbaalenii_PYR-1 IIVPPGYTDGVKFAD-----GNPYGVG-HITGPENRNELDEATLAALDHL
TH_0039|M.thermoresistible__bu LGLVAGWNSSAASEQDLNRLGFFIGAGGQTDVDAGPEQVHDADIRTCRHL
: : .*:.. : * *.. . :..: : **
MMAR_1713|M.marinum_M ARRVVHTADRLASG------------------
MUL_1950|M.ulcerans_Agy99 ARRVVHTADRLASG------------------
MAV_3849|M.avium_104 ARRVVETAGRLAS-------------------
MSMEG_2375|M.smegmatis_MC2_155 ARRVVTVAGRLAG-------------------
Mvan_2126|M.vanbaalenii_PYR-1 ARRVVGVADRLG--------------------
TH_0039|M.thermoresistible__bu GRRVATVTAQLLAGRAVTAAAADPATASAAAR
.***. .: :*