For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
NSAGESNGTRVLLVYYSYTGQTQKVLDAAADVLRERGCEVTLAPIEFVDPQYSEPFTRFPMRSVWPDMFS VYKAQGRGETGEIRTPDAVRDGRYDLVIFGSPTWWDTVSMPLRSFLESGDAKALLDGTPYAVVTVCRRLW RGNLEAVRELADKQGGRFADSIHFEYPGGQLTSLLSLASYLGSGEYKQRYLGVKIPTTNISAGQVDQARS FAGRLADRLFGGR*
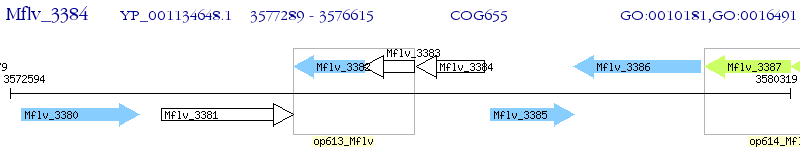
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_3384 | - | - | 100% (224) | hypothetical protein Mflv_3384 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | MAV_3849 | wrbA | 2e-05 | 28.65% (171) | NAD(P)H:quinone oxidoreductase, type IV |
| M. smegmatis MC2 155 | MSMEG_2375 | wrbA | 7e-05 | 28.90% (173) | NAD(P)H:quinone oxidoreductase, type IV |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_3117 | - | 8e-82 | 63.51% (222) | hypothetical protein Mvan_3117 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3384|M.gilvum_PYR-GCK MNSAGESNGTRVLLVYYSYTGQTQKVLDAAADVLRERGCEVTLAPIEFVD
Mvan_3117|M.vanbaalenii_PYR-1 MPTVTHSGPPRVLLLYYSYTGQSKKVLDAAADVFRARGCVVTEAPIEFTD
MAV_3849|M.avium_104 --------MTKLAIIYYSATGHGTTMARRVAAAAESAGAQVRLR--HVAE
MSMEG_2375|M.smegmatis_MC2_155 ----MTSSATKVAVIYYSATGHGTTMAQRVAASAEAAGAEVRLR--HIAE
.:: ::*** **: .: .* . *. * ...:
Mflv_3384|M.gilvum_PYR-GCK PQYSEPFTRFPMRSVWPDMFSVYKAQGRGETGEIRTPDAVRDGRYDLVIF
Mvan_3117|M.vanbaalenii_PYR-1 PRYAQRFSRFPMHRVWPEFFGMLPAQTLQRTGDIRTPDAVRTGDYDLICI
MAV_3849|M.avium_104 TQDPESFAHNP---AWTANYQATKDLPAATGDDIVWADAV--------IF
MSMEG_2375|M.smegmatis_MC2_155 TQDPQTFTQNP---AWTANYEATKDLPAATGEDIVWADAV--------VF
.: .: *:: * .*. : :* .*** :
Mflv_3384|M.gilvum_PYR-GCK GSPTWWDTVSMPLRSFLESGD---AKALLDGTPYAVVTVCRRLWRGNLEA
Mvan_3117|M.vanbaalenii_PYR-1 GSPTWWDTVSMPLRSFLKSHE---ARNLLDGKRFAVFVVCRRKWRKNLAG
MAV_3849|M.avium_104 GSPTRFGSPAAQLRAFLDSLGGLWAQGKLADKVYAAFTSSNTLHGGQETT
MSMEG_2375|M.smegmatis_MC2_155 GSPTRFGSPAAQFRAFIDSLGGLWAQGRLADKVYAAFTSSQTAHGGQEST
**** :.: : :*:*:.* *: * .. :*... .. :
Mflv_3384|M.gilvum_PYR-GCK VRELADKQGGRFADSIHFEYPGGQLTS--LLSLASYLGSGEYKQRYLGVK
Mvan_3117|M.vanbaalenii_PYR-1 VRKLAENKGGRYVDGIHFTYPGGELSS--MLSLTSYLGSGEYKDRYLGVK
MAV_3849|M.avium_104 LLSLY----------ITLMHFGGIIVAPGYTDPSKFVDGNPYGASLVTTH
MSMEG_2375|M.smegmatis_MC2_155 LLNLY----------VTLMHFGGIIVPPGYTDEVKFADGNPYGVGLIANR
: .* : : : ** : . . .: ... * : :
Mflv_3384|M.gilvum_PYR-GCK IPTTNISAGQVDQARSFAGRLADRLFGGR----
Mvan_3117|M.vanbaalenii_PYR-1 LPPTNINDEHLEESRRFAARVADKVFGRQPQGN
MAV_3849|M.avium_104 DNIHEFDEPTANALDHLARRVVETAGRLAS---
MSMEG_2375|M.smegmatis_MC2_155 TNITEIDEITNNALDHLARRVVTVAGRLAG---
::. : :* *:.