For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MGLLDDKVAFVTGAARGQGRSHAVRLAREGVSVIAVDICDEVSPDNAYPPATEADLKETAGLLEREGRPF VVDTADVRDLKALERIVTEGVERLGGRLDVVVANAGIANWARLWEMSEEQWLTMLDVNLSGVWRTMKATV PHMISAGNGGSIVLISSVAGIKSLPGQAHYSAAKHGVVGLTKSAAIELGEYGIRVNSVHPWGVATPMGVP PTEFLTEHPNYVVSFGSVLPPYAVAEPDDISDAVLWLASDLARTVTGTQLTVDMGATKV
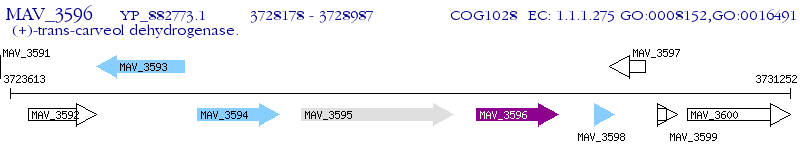
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_3596 | fabG | - | 100% (269) | 3-ketoacyl-(acyl-carrier-protein) reductase |
| M. avium 104 | MAV_4486 | - | 7e-82 | 55.93% (270) | short chain dehydrogenase |
| M. avium 104 | MAV_0846 | - | 6e-70 | 52.54% (276) | carveol dehydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0706 | fabG | 1e-82 | 56.30% (270) | 3-ketoacyl-(acyl-carrier-protein) reductase |
| M. gilvum PYR-GCK | Mflv_5072 | fabG | 2e-75 | 48.56% (278) | 3-ketoacyl-(acyl-carrier-protein) reductase |
| M. tuberculosis H37Rv | Rv0687 | fabG | 2e-82 | 56.30% (270) | 3-ketoacyl-(acyl-carrier-protein) reductase |
| M. leprae Br4923 | MLBr_02565 | fabG | 2e-25 | 33.84% (263) | 3-ketoacyl-(acyl-carrier-protein) reductase |
| M. abscessus ATCC 19977 | MAB_3844c | - | 8e-80 | 55.47% (265) | putative short-chain dehydrogenase/reductase |
| M. marinum M | MMAR_1016 | - | 5e-82 | 56.30% (270) | short-chain dehydrogenase/reductase |
| M. smegmatis MC2 155 | MSMEG_1410 | fabG | 2e-78 | 51.62% (277) | 3-ketoacyl-(acyl-carrier-protein) reductase |
| M. thermoresistible (build 8) | TH_3434 | - | 6e-82 | 54.01% (274) | PUTATIVE short-chain dehydrogenase/reductase SDR |
| M. ulcerans Agy99 | MUL_0768 | - | 1e-70 | 53.23% (248) | 3-ketoacyl-(acyl-carrier-protein) reductase |
| M. vanbaalenii PYR-1 | Mvan_1286 | fabG | 8e-77 | 50.72% (278) | 3-ketoacyl-(acyl-carrier-protein) reductase |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_1410|M.smegmatis_MC2_155 --------------------------------------------------
TH_3434|M.thermoresistible__bu --------------------------------------------------
Mflv_5072|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_1286|M.vanbaalenii_PYR-1 --------------------------------------------------
Mb0706|M.bovis_AF2122/97 --------------------------------------------------
Rv0687|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_1016|M.marinum_M --------------------------------------------------
MUL_0768|M.ulcerans_Agy99 --------------------------------------------------
MAB_3844c|M.abscessus_ATCC_199 --------------------------------------------------
MAV_3596|M.avium_104 --------------------------------------------------
MLBr_02565|M.leprae_Br4923 MAPKCSSDLFSQIVSSGPGSFVAKQLGVPRPETLRRYRPSDAPLCGSLLI
MSMEG_1410|M.smegmatis_MC2_155 --------------------------------------------------
TH_3434|M.thermoresistible__bu --------------------------------------------------
Mflv_5072|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_1286|M.vanbaalenii_PYR-1 --------------------------------------------------
Mb0706|M.bovis_AF2122/97 --------------------------------------------------
Rv0687|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_1016|M.marinum_M --------------------------------------------------
MUL_0768|M.ulcerans_Agy99 --------------------------------------------------
MAB_3844c|M.abscessus_ATCC_199 --------------------------------------------------
MAV_3596|M.avium_104 --------------------------------------------------
MLBr_02565|M.leprae_Br4923 GGDGRLVEPLRATLEKDYDLVSNNLGGRWTDSFGGLVFDATGITAPAELK
MSMEG_1410|M.smegmatis_MC2_155 --------------------------------------------------
TH_3434|M.thermoresistible__bu --------------------------------------------------
Mflv_5072|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_1286|M.vanbaalenii_PYR-1 --------------------------------------------------
Mb0706|M.bovis_AF2122/97 --------------------------------------------------
Rv0687|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_1016|M.marinum_M --------------------------------------------------
MUL_0768|M.ulcerans_Agy99 --------------------------------------------------
MAB_3844c|M.abscessus_ATCC_199 --------------------------------------------------
MAV_3596|M.avium_104 --------------------------------------------------
MLBr_02565|M.leprae_Br4923 GLHNFFTPLLRNLGRCARVVVIGTTPDATTSTDERIAQRALEGFSRSLGK
MSMEG_1410|M.smegmatis_MC2_155 -------------------------------MATVGKTLEGRVAFVTGAA
TH_3434|M.thermoresistible__bu ------------------------------------------VAFITGAA
Mflv_5072|M.gilvum_PYR-GCK ------------------------------MPMSSGTPLEGRVALITGAA
Mvan_1286|M.vanbaalenii_PYR-1 ------------------------------MPMASAAPLEGRVALITGAA
Mb0706|M.bovis_AF2122/97 -------------------------------MSARGGSLHGRVAFVTGAA
Rv0687|M.tuberculosis_H37Rv -------------------------------MSARGGSLHGRVAFVTGAA
MMAR_1016|M.marinum_M -------------------------------MAAQTGSLDGRVAFVTGAA
MUL_0768|M.ulcerans_Agy99 --------------------------------------------------
MAB_3844c|M.abscessus_ATCC_199 ---------------------------------MADQPLAGKVAFVTGAA
MAV_3596|M.avium_104 -----------------------------------MGLLDDKVAFVTGAA
MLBr_02565|M.leprae_Br4923 ELRRGATVALVYLSPDAKPAATGLESTMRFILSAKSAYVNGQVFYVGAAD
MSMEG_1410|M.smegmatis_MC2_155 RGQGR------AHAVRLADEGADIIAIDVCAPVDDTITYPSATPEDLAET
TH_3434|M.thermoresistible__bu RGQGR------AHAIRLAEEGADIIAIDVCGPVSDTITYPPATEDDLAET
Mflv_5072|M.gilvum_PYR-GCK RGQGR------AHAIRLAADGADIVALDVCKPVSESVTYPMPTSEDLAET
Mvan_1286|M.vanbaalenii_PYR-1 RGQGR------AHAVRLAAEGADIIAIDVCGPVSESITYSMPTSEDLAET
Mb0706|M.bovis_AF2122/97 RAQGR------SHAVRLAREGADIVALDICAPVSGSVTYPPATSEDLGET
Rv0687|M.tuberculosis_H37Rv RAQGR------SHAVRLAREGADIVALDICAPVSGSVTYPPATSEDLGET
MMAR_1016|M.marinum_M RAQGR------SHAVRLAREGADIIAMDICGPVSETITYAPATAADLSET
MUL_0768|M.ulcerans_Agy99 --------------MRLAREGADIIAMDICGPVSESITYAPATAADLSET
MAB_3844c|M.abscessus_ATCC_199 RGQGR------QHAVRLAQAGADVVAIDACAPVSEYAGYEAATPEDLKET
MAV_3596|M.avium_104 RGQGR------SHAVRLAREGVSVIAVDICDEVSPDNAYPPATEADLKET
MLBr_02565|M.leprae_Br4923 STPPTNWDRPLENKVAIVTGAARGIGAAIAEVVARDGARVVAIDVESAAE
: :. .. :. . * . :
MSMEG_1410|M.smegmatis_MC2_155 VAAVEATGRKVLAREVDIRDLAAQQQVVADGVEQFG-RLDIVVANAGVLS
TH_3434|M.thermoresistible__bu VRAVEATGRKVLARTADIRDLAAQQQIVAEGMERFG-RLDIVVANAGVLS
Mflv_5072|M.gilvum_PYR-GCK VRLVEETGRKVLAREVDIRDLAAQQQLVADAVEQFG-RLDIVVANAGVLS
Mvan_1286|M.vanbaalenii_PYR-1 VRLVEATGRKVLAREVDIRDLAAQQQVVADGVEQFG-RLDIVVANAGVLS
Mb0706|M.bovis_AF2122/97 VRAVEAEGRKVLAREVDIRDDAELRRLVADGVEQFG-RLDIVVANAGVLG
Rv0687|M.tuberculosis_H37Rv VRAVEAEGRKVLAREVDIRDDAELRRLVADGVEQFG-RLDIVVANAGVLG
MMAR_1016|M.marinum_M IRAVESEGRKVLARQADVRDSAALQQLVADGVEEFG-RLDVVVANAGVLG
MUL_0768|M.ulcerans_Agy99 IRAVESEGRKVLARQADVRDSAALQQLVADGVEEFG-RLDVMVANAGVFG
MAB_3844c|M.abscessus_ATCC_199 VRLVEAAGRKIIAEQADVRNGAALQSVVEEAVAQFG-HIDILIANAGVLS
MAV_3596|M.avium_104 AGLLEREGRPFVVDTADVRDLKALERIVTEGVERLGGRLDVVVANAGIAN
MLBr_02565|M.leprae_Br4923 ALDETANRVGGTTLSLDVTADDAVDKITEHLHDYQGGHADILVNNAGITR
. *: :. . * : *::: ***:
MSMEG_1410|M.smegmatis_MC2_155 WGRIFEMSPEQWDTVVDVNLNGTWRTIRAAVPAMIEAGNGGSIIIVSSSA
TH_3434|M.thermoresistible__bu WGRLWEMSEEQWDTVIDVNLNGTWRTIRAVVPAMIEAGNGGSIIIVSSSA
Mflv_5072|M.gilvum_PYR-GCK WGRIWEMSEDQWDTVVDVNLNGTWRTIRAAVPAMIEAGNGGSIIIVSSSA
Mvan_1286|M.vanbaalenii_PYR-1 WGRIWEMSEEQWDTVVDVNLNGTWRTVRAAVPAMIEAGNGGSIVIVSSSA
Mb0706|M.bovis_AF2122/97 WGRLWELTDEQWETVIGVNLTGTWRTLRATVPAMIDAGNGGSIVVVSSSA
Rv0687|M.tuberculosis_H37Rv WGRLWELTDEQWETVIGVNLTGTWRTLRATVPAMIDAGNGGSIVVVSSSA
MMAR_1016|M.marinum_M WGRLWELTDEQWDTVIGVNLSGTWRTLRAAVPAMIEAGNGGSIIVVSSSA
MUL_0768|M.ulcerans_Agy99 WGRLWELTDEQWDTVIGVNLSGTWRTLRAAVPAMIEAGNGGSIIVVSSSA
MAB_3844c|M.abscessus_ATCC_199 WGRLWEMSDQQWEDIIDTNLTGTWKTVKAVVPTMIEAGNGGSIVIVSSVS
MAV_3596|M.avium_104 WARLWEMSEEQWLTMLDVNLSGVWRTMKATVPHMISAGNGGSIVLISSVA
MLBr_02565|M.leprae_Br4923 DKLLANMDDCRWDAVLEVNLLAPQRLTEGLVGNGSIS-KGGRVIVLSSMA
: :: :* :: .** . : .. * : :** ::::** :
MSMEG_1410|M.smegmatis_MC2_155 GLKATPGNGHYAASKHGLVALTNSLALEVGEYGIRVNSIHPYSIDTPMIE
TH_3434|M.thermoresistible__bu GLKATPGNGHYAASKHGLVALTNALAIEVGEYGIRVNSIHPYSIDTPMIE
Mflv_5072|M.gilvum_PYR-GCK GLKATPGNAHYAASKHGLVALTNALALEAGEFGIRVNSIHPYSIDTPMIE
Mvan_1286|M.vanbaalenii_PYR-1 GLKATPGNAHYAASKHGLVALTNALAIEAGEFGIRVNSIHPYSIDTPMIE
Mb0706|M.bovis_AF2122/97 GLKATPGNGHYAASKHALVALTNTLAIELGEFGIRVNSIHPYSVDTPMIE
Rv0687|M.tuberculosis_H37Rv GLKATPGNGHYAASKHALVALTNTLAIELGEFGIRVNSIHPYSVDTPMIE
MMAR_1016|M.marinum_M GLKATPGNGHYAASKHGLVALTNTLAIELGEYDIRVNSIHPYSVETPMIE
MUL_0768|M.ulcerans_Agy99 GLKATPGNGHYAASKHGLVALTNTLAIELGEYDIGVNSIHPYSVETPMIE
MAB_3844c|M.abscessus_ATCC_199 GLKGTPGNGAYVASKFGLTGLTKSLTIELAEYGIRVNSIHPYGVTTPMIT
MAV_3596|M.avium_104 GIKSLPGQAHYSAAKHGVVGLTKSAAIELGEYGIRVNSVHPWGVATPMGV
MLBr_02565|M.leprae_Br4923 GIAGNRGQTNYATAKAGVIGLTQALAPGLAEKGITINAVAPGFIETQMTA
*: . *: * ::* .: .**:: : .* .* :*:: * : * *
MSMEG_1410|M.smegmatis_MC2_155 PEAMAQVFAKYPNYLHSFAPMPYQPVG-EGAEGLSTFMTPEEVSDVVAWL
TH_3434|M.thermoresistible__bu RDAMMAVFAKHPNYLHSFAPMPLHPVNREGDGGRQEFMTPEEVADVVAWL
Mflv_5072|M.gilvum_PYR-GCK KDAMMEIFSKYPTFLHSFAPMPFKPVARDGKPGLQEFMTVEEVSDVVAWL
Mvan_1286|M.vanbaalenii_PYR-1 KDAMMEIFSKYPTYLHSFAPMPFKPVAREGKPGLQEFMTVEEVADVVAWL
Mb0706|M.bovis_AF2122/97 PEAMIQTFAKHPGYVHSFPPMPLQPKG---------FMTPDEISDVVVWL
Rv0687|M.tuberculosis_H37Rv PEAMIQTFAKHPGYVHSFPPMPLQPKG---------FMTPDEISDVVVWL
MMAR_1016|M.marinum_M PDVMMQVFGEHPRFLHSFPPMPLQYKG---------LMTSEEVSDVVVWL
MUL_0768|M.ulcerans_Agy99 PDVMMQVFGEHPRFLHSFPPMPLQYKG---------LMTSEEVSDVVVWL
MAB_3844c|M.abscessus_ATCC_199 GDAMMKILAEHPNYLPSIAPMPLSPTK---------MLEPDEISDVVLWL
MAV_3596|M.avium_104 PP--TEFLTEHPNYVVSFG-SVLPPYA---------VAEPDDISDAVLWL
MLBr_02565|M.leprae_Br4923 AIPLATREVGRR--LNSLLQGG----------------LPVDVAETIAYF
: *: ::::.: ::
MSMEG_1410|M.smegmatis_MC2_155 AGDGSATLSGSQIAVDRGVMKY-
TH_3434|M.thermoresistible__bu AGDGSATLSGSQIAVDRGVMKY-
Mflv_5072|M.gilvum_PYR-GCK AGDSSATISGSQIAVDRGTMKY-
Mvan_1286|M.vanbaalenii_PYR-1 AGDASSTISGSQIAVDRGTMKY-
Mb0706|M.bovis_AF2122/97 AGDGSGALSGNQIPVDKGALKY-
Rv0687|M.tuberculosis_H37Rv AGDGSGALSGNQIPVDKGALKY-
MMAR_1016|M.marinum_M AGDGSGTLSGAQIPVDKGALKY-
MUL_0768|M.ulcerans_Agy99 AGDGSGTLSGAQIPVDKGALKY-
MAB_3844c|M.abscessus_ATCC_199 ASDASGTLAGAQVPVDKGHLTY-
MAV_3596|M.avium_104 ASDLARTVTGTQLTVDMGATKV-
MLBr_02565|M.leprae_Br4923 AIPASNAVTGNVIRVCGQAMLGA
* : :::* : *