For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
IRYSTQPRRLRVSALAAVANPSYARVDTWNLLDDACRHLAEVDLAGLDKTHDVARVKRLLDRIAAYERYW LYPGAENLAIFRAHLESLSTVRLTEEVSLAVRLLSEYGDRAGLFDTSAPLDDQELVAQAKQQHFYTVLLA DDAPSTAPDSLAECLRALRNPSDDVQFEILVVPSVEDAITAVALNGEIQAAIIRDDLPLRSRDRLPLMNT LLGPNEDADGVIPDRANDWVECGEWIRELRPHIDLYLLTDESIAAGDDTEPDVYDRTFYRLNDVTDLHST VLAGLRNRYATPFFDALRAYAAAPVGQFHALPVARGASIFNSRSLQDMGEFYGRNIFMAETSTTSGGLDS LLDPHGNIKKAMDKAAKTWNADHTYFVTNGTSTANKIVVQSLTRPGDIVLIDRNCHKSHHYGLVLAGAYP LYLDAYPLPQFAIYGAVSLRTIKQTLLDLEAAGQLHRVRMLLLTNCTFDGVVYNPLQVMQEVLAIKPDIC FLWDEAWYAFATAVPWARQRTAMVAAERLEQMLASPDYVEQYRKWAASMQGVDRSEWIERELLPDPATAR VRVYATHSTHKSLSALRQASMIHVRDQDFNALTRDAFGEAFLTHTSTSPNQQLLASLDLARRQVDIEGFQ LVRQVYDMALVFRHRVRKDRLISKWFRILDESDLVPEEFRASSVSSYREVRQGALAEWNEAWRSDQFVLD ATRVTLFVGATGMNGYDFREKILMERFGIQINKTSINSVLLIFTIGVTWSSVHYLLDVLRRVATDFDRIQ KEASAADRALQRRHVEEITEDLPHLPDFSEFDVAFRPVDECNFGDMRSAFYAGYEESDREHVLIGTAGRR LAEGKNLVSTTFVVPYPPGFPVLVPGQVVSKEIVYFLAQLDVKEIHGYNPDLGLSVFTDTALARMEAQRN AATAAVGSVTAAFELPADASGGNGAHNGAGAVPSVADGG*
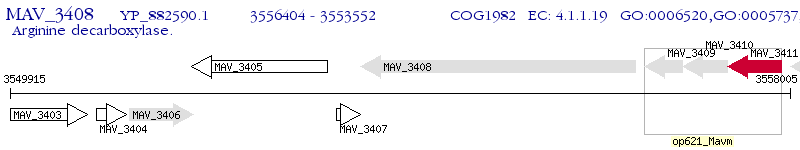
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_3408 | - | - | 100% (950) | putative Orn/Lys/Arg decarboxylase |
| M. avium 104 | MAV_2031 | - | 3e-08 | 24.28% (313) | Orn/Lys/Arg decarboxylase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2560c | - | 0.0 | 85.91% (944) | amino acid decarboxylase |
| M. gilvum PYR-GCK | - | - | - | - | - |
| M. tuberculosis H37Rv | Rv2531c | - | 0.0 | 85.91% (944) | amino acid decarboxylase |
| M. leprae Br4923 | MLBr_00524 | adi | 0.0 | 86.60% (940) | putative amino acid decarboxylase |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | MMAR_2184 | - | 0.0 | 86.81% (948) | amino acid decarboxylase |
| M. smegmatis MC2 155 | MSMEG_0266 | - | 2e-07 | 21.54% (311) | arginine decarboxylase |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_2916 | - | 5e-14 | 26.59% (346) | Orn/Lys/Arg decarboxylase, major region |
CLUSTAL 2.0.9 multiple sequence alignment
MAV_3408|M.avium_104 MIRYSTQPRRLRVSALAAVANPSYARVDTWNLLDDACRHLAEVDLAGLDK
MMAR_2184|M.marinum_M MKRYSTRPRRLRVSALAAVANPSYARVDTWNLLDDACCHLAEVDLAGMDI
Mb2560c|M.bovis_AF2122/97 MNPNSVRPRRLHVSALAAVANPSYTRLDTWNLLDDACRHLAEVDLAGLDT
Rv2531c|M.tuberculosis_H37Rv MNPNSVRPRRLHVSALAAVANPSYTRLDTWNLLDDACRHLAEVDLAGLDT
MLBr_00524|M.leprae_Br4923 MNRDSGHPRRLRVSALAAVANPSYTRIDTWNVLDDACRHLAEVDRAGLDT
Mvan_2916|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_0266|M.smegmatis_MC2_155 --------------------------------------------------
MAV_3408|M.avium_104 THDVARVKRLLDRIAAYERYWLYPGAENLAIFRAHLESLSTVRLTEEVSL
MMAR_2184|M.marinum_M THDMAKVRRLMDRIGAYERYWLYPGAQNLANFRAHLESKSTVRLKEEVSL
Mb2560c|M.bovis_AF2122/97 THDVARAKRLMDRIGAYERYWLYPGAQNLATFRAHLDSHSTVRLTEEVSL
Rv2531c|M.tuberculosis_H37Rv THDVARAKRLMDRIGAYERYWLYPGAQNLATFRAHLDSHSTVRLTEEVSL
MLBr_00524|M.leprae_Br4923 AHDVARVKRLLDRIGAYERYWLYPGAANLAAFRGYLATMATVQLTEEVSL
Mvan_2916|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_0266|M.smegmatis_MC2_155 --------------------------------------------------
MAV_3408|M.avium_104 AVRLLSEYGDRAGLFDTSAPLDDQELVAQAKQQHFYTVLLADDAPSTAPD
MMAR_2184|M.marinum_M AVRLLGEYGDRTALFDTSAPLAEQELVAQAKQQQFYTVLVADDAPTSAPD
Mb2560c|M.bovis_AF2122/97 AVRLLSEYGDRTALFDTSASLAEQELVAQAKQQQFYTVLLADDSPATAPD
Rv2531c|M.tuberculosis_H37Rv AVRLLSEYGDRTALFDTSASLAEQELVAQAKQQQFYTVLLADDSPATAPD
MLBr_00524|M.leprae_Br4923 AVRLLSEYGDRTALFDTSAPLADQELVAQAKQQQFYTVLLADDAPSTAPD
Mvan_2916|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_0266|M.smegmatis_MC2_155 --------------------------------------------------
MAV_3408|M.avium_104 SLAECLRALRNPSDDVQFEILVVPSVEDAITAVALNGEIQAAIIRDDLPL
MMAR_2184|M.marinum_M SLAEHLRALHNPSDDVQFEVLLAPSVEDAITAVALNGEIQAAIIRHDLPL
Mb2560c|M.bovis_AF2122/97 SLAECLRQLRNPADEVQFELLVVASIEDAITAVALNGEIQAAIIRHDLPL
Rv2531c|M.tuberculosis_H37Rv SLAECLRQLRNPADEVQFELLVVASIEDAITAVALNGEIQAAIIRHDLPL
MLBr_00524|M.leprae_Br4923 CLAESLRELRNPSDDVQFELLVVSSVEDAITAVALNGEIQAAIIRHDLPL
Mvan_2916|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_0266|M.smegmatis_MC2_155 --------------------------------------------------
MAV_3408|M.avium_104 RSRDRLPLMNTLLGPNEDADG-VIPDRANDWVECGEWIRELRPHIDLYLL
MMAR_2184|M.marinum_M RSRDRLPLMNALLGAN--DDV-VAPDSPHDWIECGEWIRELRPHIDLYLL
Mb2560c|M.bovis_AF2122/97 RSRDRVPLMTTLLGTDGDE---AVANETHDWVECAEWIRELRPHIDLYLL
Rv2531c|M.tuberculosis_H37Rv RSRDRVPLMTTLLGTDGDE---AVANETHDWVECAEWIRELRPHIDLYLL
MLBr_00524|M.leprae_Br4923 RSRDRVPLMNTLLGANDSEGALVTIDRPHDWVECGEWIRELRPHIDLYLL
Mvan_2916|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_0266|M.smegmatis_MC2_155 --------------------------------------------------
MAV_3408|M.avium_104 TDESIAAGDDTEPDVYDRTFYRLNDVTDLHSTVLAGLRNRYATPFFDALR
MMAR_2184|M.marinum_M TDESIAAGAEDDPDVYDRTFYRLNDVTDLHSTVLAGLRNRFATPFFDALR
Mb2560c|M.bovis_AF2122/97 TDESIAAETQDEPDVYDRTFYRLNDVTDLHSTVLAGLRNRYATPFFDALR
Rv2531c|M.tuberculosis_H37Rv TDESIAAETQDEPDVYDRTFYRLNDVTDLHSTVLAGLRNRYATPFFDALR
MLBr_00524|M.leprae_Br4923 TDESIAAGNDDEPDVYDRTFYRLNDVTDLNSTVLAGLRNRFATPFFDALR
Mvan_2916|M.vanbaalenii_PYR-1 ----------------------------------MGFEHS-RAPLFESLR
MSMEG_0266|M.smegmatis_MC2_155 -------------------------------------MDQTETPLLDALN
. :*::::*.
MAV_3408|M.avium_104 AYAAAPVGQFHALPVARGASIFNSRSLQDMGEFYGRNIFMAETS---TTS
MMAR_2184|M.marinum_M AYAAAPVGQFHALPVARGASIFNSKSLQDMGEFYGRNIFMAETS---TTS
Mb2560c|M.bovis_AF2122/97 AYAAAPVGQFHALPVARGASIFNSKSLHDMGEFYGRNIFMAETS---TTS
Rv2531c|M.tuberculosis_H37Rv AYAAAPVGQFHALPVARGASIFNSKSLHDMGEFYGRNIFMAETS---TTS
MLBr_00524|M.leprae_Br4923 AYAAAPVGQFHALPVARGASIFNSRSLQDMGEFYGRNIFMAETS---TTS
Mvan_2916|M.vanbaalenii_PYR-1 TFVERDQAPFYSP---------GHKGGRTIDPWLRENIAAVDLN---NLP
MSMEG_0266|M.smegmatis_MC2_155 EYHASNRYGYSPP---------GHRQGRGVDDRVLRVLGREPFLSDVLAN
: : . . : : :. . :
MAV_3408|M.avium_104 GGLDSLLDPHGNIKKAMDKAAKTWNADHTYFVTNGTSTANKIVVQSLTRP
MMAR_2184|M.marinum_M GGLDSLLDPHGNIKKAMDKAASTWNADHTYFVTNGTSTANKIVVQSLTRP
Mb2560c|M.bovis_AF2122/97 GGLDSLLDPHGNIKTAMDKAAVTWNANQTYFVTNGTSTANKIVVQALTRP
Rv2531c|M.tuberculosis_H37Rv GGLDSLLDPHGNIKTAMDKAAVTWNANQTYFVTNGTSTANKIVVQALTRP
MLBr_00524|M.leprae_Br4923 GGLDSLLDPHGNIKKAMDKAAVTWRANHTYFVTNGTSTANKIVVQSLTRP
Mvan_2916|M.vanbaalenii_PYR-1 -DTDTLHCPEGPILEAERLIADAWGVPQSFLMVQGSTGGNIAVALTALRP
MSMEG_0266|M.smegmatis_MC2_155 GGLDDRRSSNGYLQKAEDLMAEAVGADTAWFSTCGSSLSVKAAMMAVAGG
. * ..* : * * : . ::: . *:: . . :
MAV_3408|M.avium_104 GDIVLIDRNCHKSHHYGLVLAGAYPLYLDAYPLPQFAIYGAVSLRTIKQT
MMAR_2184|M.marinum_M GDIVLIDRNCHKSHHYGLVLAGAYPLYLDAYPLPPFAIYGAVSLHTIKQV
Mb2560c|M.bovis_AF2122/97 GDIVLIDRNCHKSHHYGLVLAGAYPMYLDAYPLPQYAIYGAVPLRTIKQA
Rv2531c|M.tuberculosis_H37Rv GDIVLIDRNCHKSHHYGLVLAGAYPMYLDAYPLPQYAIYGAVPLRTIKQA
MLBr_00524|M.leprae_Br4923 GDIVLIDRNCHKSHHYGLVLAGAYPMYLDAYPLPQFAIYGAVSLRTIKKA
Mvan_2916|M.vanbaalenii_PYR-1 DEPVLVARNAHKSVLAGLVQVGARPVWLEPRWDAEFGVAHGLDAAVVARA
MSMEG_0266|M.smegmatis_MC2_155 DGSLLLSRDSHKSVVAGLIFSGVMPRWITPRWDADRHLSHPPSPEEVEET
. :*: *:.*** **: *. * :: . . : : ..
MAV_3408|M.avium_104 LLDLEAAGQLHRVRMLLLTNCTFDGVVYNPLQVMQEVLAIKPDICFLWDE
MMAR_2184|M.marinum_M LLDLEAAGQLHKVRMLLLTNCTFDGVVYNPRLVMEEVLAIKPDICFLWDE
Mb2560c|M.bovis_AF2122/97 LLDLEAAGQLHRVRMLLLTNCTFDGVVYNPRRVMEEVLAIKPDICFLWDE
Rv2531c|M.tuberculosis_H37Rv LLDLEAAGQLHRVRMLLLTNCTFDGVVYNPRRVMEEVLAIKPDICFLWDE
MLBr_00524|M.leprae_Br4923 LLDLEAVRQLDRVRMLLLTNCTFDGVVCNPQRVMEEVLAIKPDICFLWDE
Mvan_2916|M.vanbaalenii_PYR-1 FETTGAT-------ALWALHPTYYGTTGD--------LAALADLCRRYGA
MSMEG_0266|M.smegmatis_MC2_155 WRRYPDAAG------ALIVSPSPYGTCAD--------LAGIADVCHARGK
. : *. : ** .*:* .
MAV_3408|M.avium_104 AWYAFATAVPWARQRTAMVAAERLEQMLASPDYVEQYRKWAASMQGVDRS
MMAR_2184|M.marinum_M AWYAFATAVPWARQRTAMVSAEYLESMLASDEYGAVYEKWRASMQGIPRS
Mb2560c|M.bovis_AF2122/97 AWYAFATAVPWARQRTAMIAAERLEQMLSTAEYAEEYRNWCASMDGVDRS
Rv2531c|M.tuberculosis_H37Rv AWYAFATAVPWARQRTAMIAAERLEQMLSTAEYAEEYRNWCASMDGVDRS
MLBr_00524|M.leprae_Br4923 AWYAFATAVPWARQRTAMVSAEQLEQKLWSVEYAEEYRNWCASMAEIPRS
Mvan_2916|M.vanbaalenii_PYR-1 R----------------------------------------LLLDGAHSP
MSMEG_0266|M.smegmatis_MC2_155 P----------------------------------------LIVDEAWGA
: .
MAV_3408|M.avium_104 EWIERELLPDPATARVRVYATHSTHKSLSALRQASMIHVRDQDFNALTRD
MMAR_2184|M.marinum_M QWAEHRLLPDPKRARVRVYATHSTHKSLSALRQASMIHVRDQDFNALTRD
Mb2560c|M.bovis_AF2122/97 EWVDHRLLPDPNRARVRVYATHSTHKSLSALRQASMIHVRDQDFKALTRD
Rv2531c|M.tuberculosis_H37Rv EWVDHRLLPDPNRARVRVYATHSTHKSLSALRQASMIHVRDQDFKALTRD
MLBr_00524|M.leprae_Br4923 QWVDQRLLPDPNRARIRVYATHSTHKSLSALRQASMIHVRDQDFNALARD
Mvan_2916|M.vanbaalenii_PYR-1 HFAFHPDLPTPGEQSGAAATVHSVHKILSGLSQAAVLHVDTAQLDEAS--
MSMEG_0266|M.smegmatis_MC2_155 HLPFHENLPTWAMNAGADVCVVSVHKMGAGFEQGSAFHLQG---DLIDQD
. : ** . *.** :.: *.: :*: .
MAV_3408|M.avium_104 AFGEAFLTHTSTSPNQQLLASLDLARRQVDIEGFQLVRQVYDMALVFRHR
MMAR_2184|M.marinum_M AFGEAFLTHTSTSPNQQLLASLDLARRQVDIEGFQLVRQVYDMALVFRHR
Mb2560c|M.bovis_AF2122/97 AFGEAFLTHTSTSPNQQLLASLDLARRQVDIEGFELVRHVYNMALVFRHR
Rv2531c|M.tuberculosis_H37Rv AFGEAFLTHTSTSPNQQLLASLDLARRQVDIEGFELVRHVYNMALVFRHR
MLBr_00524|M.leprae_Br4923 AFGEAFLTHTSTSPNQQLLASLDLARRQVDIEGFQLVRLVYDMALVFRHR
Mvan_2916|M.vanbaalenii_PYR-1 -VRRSLQLIQTTSPHFAIMASVDLARRQMVLDGHALLDATLTRAREAADR
MSMEG_0266|M.smegmatis_MC2_155 RLSACADLLMTTSPNVLVYSAIDGWRRQMVEHGHRLFGAALDLADDTRRR
. . :***: : :::* ***: .*. *. . * *
MAV_3408|M.avium_104 VRKDRLISKWFRILDESDLVPEEFRASSVSSYREVRQGALAEWNEAWRSD
MMAR_2184|M.marinum_M VRKDRLISKWFRILDEADLVPEEFRASSVSSYRQVRQGALAEWNEAWRSD
Mb2560c|M.bovis_AF2122/97 VRKDRLISKWFRILDESDLVPDAFRSSTVSSYRQVRQGALADWNEAWRSD
Rv2531c|M.tuberculosis_H37Rv VRKDRLISKWFRILDESDLVPDAFRSSTVSSYRQVRQGALADWNEAWRSD
MLBr_00524|M.leprae_Br4923 VRKDRLISKWFRILDESDLVPDEYRASAVSSYRQVRQGALAEWNEAWRSD
Mvan_2916|M.vanbaalenii_PYR-1 LS---------QVPGLTVLRPEHLAGAGTG--------------------
MSMEG_0266|M.smegmatis_MC2_155 IE----------EIDDVEVLDEELLGAQAS--------------------
: . : : .: ..
MAV_3408|M.avium_104 QFVLDATRVTLFVGATGMNGYDFREKILMERFGIQINKTSINSVLLIFTI
MMAR_2184|M.marinum_M QFVLDATRVTLFIGSTGMNGYDFREKILMERFGIQINKTSINSVLLIFTI
Mb2560c|M.bovis_AF2122/97 QFVLDPTRLTLFIGATGMNGYDFREKILMERFGIQINKTSINSVLLIFTI
Rv2531c|M.tuberculosis_H37Rv QFVLDPTRLTLFIGATGMNGYDFREKILMERFGIQINKTSINSVLLIFTI
MLBr_00524|M.leprae_Br4923 QFVLDPTRVTLFVGKTGMNGYDFREKILMERFGIQINKTSINSVLLIFTI
Mvan_2916|M.vanbaalenii_PYR-1 LCQLDETKLLVGTAGVDADARDILG-RLNRIHGVQPELAGAGHILCISTI
MSMEG_0266|M.smegmatis_MC2_155 -HDLDRLQILIDVSATGTSGYQAAD-WLRENCQVDVGMSDHRRILATMSV
** :: : . .. .. : * . :: :. :* ::
MAV_3408|M.avium_104 GVTWSSVHYLLDVLRRVATDFDRIQKEASAADRALQRRHVEEITEDLPHL
MMAR_2184|M.marinum_M GVTWSSVHYLLDVLRRIATDFDRSQRAASAADWALHERRVEEITQDLPHL
Mb2560c|M.bovis_AF2122/97 GVTWSSVHYLLDVLRRVAIDLDRSQKAASGADLALHRRHVEEITQDLPHL
Rv2531c|M.tuberculosis_H37Rv GVTWSSVHYLLDVLRRVAIDLDRSQKAASGADLALHRRHVEEITQDLPHL
MLBr_00524|M.leprae_Br4923 GVTWSSVHYLLDVLRRVASDFDRIQKVASGADLALHQRHVEEITQDLPHL
Mvan_2916|M.vanbaalenii_PYR-1 GNIDDDFYRLTTAFEEVSAHFGRRDAEGAG--------------------
MSMEG_0266|M.smegmatis_MC2_155 ADGSDTAGRLLHALSLWRKAADDFAPPPPID------------------L
. . * .: . .
MAV_3408|M.avium_104 PDFSEFDVAFRPVDECNFGDMRSAFYAGYEESDREHVLIGTAGRRLAEGK
MMAR_2184|M.marinum_M PDFSEFDIAFRPDDASSFGDMRSAFYAGYDESDREYVQIGMAGRRLAEGR
Mb2560c|M.bovis_AF2122/97 PDFSEFDLAFRPDDASSFGDMRSAFYAGYEEADREYVQIGLAGRRLAEGK
Rv2531c|M.tuberculosis_H37Rv PDFSEFDLAFRPDDASSFGDMRSAFYAGYEEADREYVQIGLAGRRLAEGK
MLBr_00524|M.leprae_Br4923 PDFSEFDVAFRPENASSFGDMRSAFYAGYEESDREYVHIGTAGRQLAEGR
Mvan_2916|M.vanbaalenii_PYR-1 -AWTADVLAVRPELVLTP---REAFFAADET------------VALADAA
MSMEG_0266|M.smegmatis_MC2_155 PSPAELQLTTKTS-------PRDAFFAEVEQ------------IPVEHAV
: :: :. *.**:* : : ..
MAV_3408|M.avium_104 NLVSTTFVVPYPPGFPVLVPGQVVSKEIVYFLAQLDVKEIHGYN-PDLGL
MMAR_2184|M.marinum_M TLVSTTFVVPYPPGFPVLVPGQVVSKEIVYFLAQLDVKEIHGYN-HELGL
Mb2560c|M.bovis_AF2122/97 TLVSTTFVVPYPPGFPVLVPGQLVSKEIIYFLAQLDVKEIHGYN-PDLGL
Rv2531c|M.tuberculosis_H37Rv TLVSTTFVVPYPPGFPVLVPGQLVSKEIIYFLAQLDVKEIHGYN-PDLGL
MLBr_00524|M.leprae_Br4923 TLVSTTFVVPYPPGFPVLVPGQVVSKEIIYFLAQLDVKEIHGYN-HELGL
Mvan_2916|M.vanbaalenii_PYR-1 GRIAAEAITPYPPGIPVVMPGERLGKDVVDLLLRLRAAGNPISA-SDPTM
MSMEG_0266|M.smegmatis_MC2_155 GRIAAEQITPYPPGIPAVVPGERLNEAVLDYLVTGVQAGMNLPDPADPQM
::: :.*****:*.::**: :.: :: * : :
MAV_3408|M.avium_104 SVFTDTALARMEAQRNAATAAVGSVTAAFELPADASGGNGAHNGAGAVPS
MMAR_2184|M.marinum_M SVFTDVALKRMEAARN-VIASAGAELPAFELAPASS----VVNGDGVLQA
Mb2560c|M.bovis_AF2122/97 SVFTQAALARMEAARN-AVATVGAALPAFEVPRDASALNGTVNGDSVLQG
Rv2531c|M.tuberculosis_H37Rv SVFTQAALARMEAARN-AVATVGAALPAFEVPRDASALNGTVNGDSVLQG
MLBr_00524|M.leprae_Br4923 SVFTEKALARMIARRNAASAAVGSAFAAFEIPSDSAAMGGDVNGD-RVEA
Mvan_2916|M.vanbaalenii_PYR-1 GVVKTVR-------------------------------------------
MSMEG_0266|M.smegmatis_MC2_155 GSIRVMA-------------------------------------------
. .
MAV_3408|M.avium_104 VADGG
MMAR_2184|M.marinum_M VADDA
Mb2560c|M.bovis_AF2122/97 VAEDA
Rv2531c|M.tuberculosis_H37Rv VAEDA
MLBr_00524|M.leprae_Br4923 VAEDA
Mvan_2916|M.vanbaalenii_PYR-1 -----
MSMEG_0266|M.smegmatis_MC2_155 -----