For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VDAESAARVGTVAASATGEVNQRDWQRWAAAVGDHNPLYFDPDYARANGFRDIICPPLFLQYAILGVSRL DSLRPDGSSGAVSGSLAFPRAPKRMAGGESFTFHLPAYHRDRVDMVRTIASIVEKQGRSGRFVLVTWHTV YRNQHDELLAEASTSMIARP*
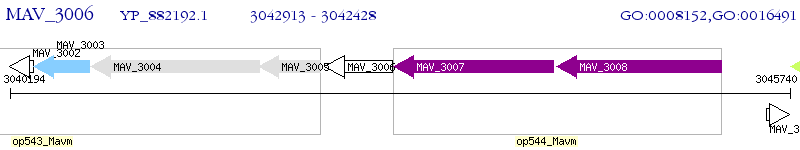
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_3006 | - | - | 100% (161) | hypothetical protein MAV_3006 |
| M. avium 104 | MAV_0619 | - | 6e-06 | 38.98% (59) | hypothetical protein MAV_0619 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3572c | - | 9e-06 | 39.34% (61) | hypothetical protein Mb3572c |
| M. gilvum PYR-GCK | Mflv_1511 | - | 2e-06 | 34.25% (73) | hypothetical protein Mflv_1511 |
| M. tuberculosis H37Rv | Rv3542c | - | 9e-06 | 39.34% (61) | hypothetical protein Rv3542c |
| M. leprae Br4923 | MLBr_02566 | - | 0.0001 | 33.87% (62) | hypothetical protein MLBr_02566 |
| M. abscessus ATCC 19977 | MAB_2054 | - | 8e-11 | 29.22% (154) | hypothetical protein MAB_2054 |
| M. marinum M | MMAR_4247 | - | 1e-10 | 32.39% (142) | hypothetical protein MMAR_4247 |
| M. smegmatis MC2 155 | MSMEG_5992 | - | 1e-05 | 40.43% (47) | hypothetical protein MSMEG_5992 |
| M. thermoresistible (build 8) | TH_4561 | - | 5e-09 | 27.59% (145) | PUTATIVE conserved hypothetical protein |
| M. ulcerans Agy99 | MUL_0938 | - | 6e-10 | 31.69% (142) | hypothetical protein MUL_0938 |
| M. vanbaalenii PYR-1 | Mvan_5255 | - | 1e-05 | 40.43% (47) | hypothetical protein Mvan_5255 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1511|M.gilvum_PYR-GCK ---------------------------------------------MSADL
MSMEG_5992|M.smegmatis_MC2_155 -----------------------------------------MTTQVNEDL
Mvan_5255|M.vanbaalenii_PYR-1 --------------------------------------MREERTIGMSDL
Mb3572c|M.bovis_AF2122/97 -------------------------------------------MTGVSDI
Rv3542c|M.tuberculosis_H37Rv -------------------------------------------MTGVSDI
MMAR_4247|M.marinum_M ---------------------------------------MTDSETEFLSK
MUL_0938|M.ulcerans_Agy99 MRRCVRSAAIREVPGGVGTATRPMTRRRSDASTSQEDNRVTDSETEFLFR
TH_4561|M.thermoresistible__bu --------------------------------------MTEISKSELRQR
MAB_2054|M.abscessus_ATCC_1997 ---------------------------------------------MTRVA
MAV_3006|M.avium_104 ---------------------------------------------MVDAE
MLBr_02566|M.leprae_Br4923 -----------------------------------------MTFGFVLRR
Mflv_1511|M.gilvum_PYR-GCK QAGIEKITAAGRSEPRAGRDPVNQPMIHHWVDAIGDRNPIYVDDEAARAA
MSMEG_5992|M.smegmatis_MC2_155 QAGIEKIKAEGKSDPRKGRDPVNQPMIHHWVDAIGDKNPIYVDQEAAKAA
Mvan_5255|M.vanbaalenii_PYR-1 QSAIAEIKAAGRSKPRAGRDPVNQPMIHHWVDAIGDKNPIYVDDEAAKAA
Mb3572c|M.bovis_AF2122/97 QEAVAQIKAAGPSKPRLARDPVNQPMINNWVEAIGDRNPIYVDDAAARAA
Rv3542c|M.tuberculosis_H37Rv QEAVAQIKAAGPSKPRLARDPVNQPMINNWVEAIGDRNPIYVDDAAARAA
MMAR_4247|M.marinum_M LRSLVGQPTGGTGRPSVAPDPVNQPMIRHWAHALDDMNPVYLDPEFAAAS
MUL_0938|M.ulcerans_Agy99 LRSLVGQPTGGTGRPSVAPDPVNQPMIRHWAHALDDMNPVYLDPEFAAAS
TH_4561|M.thermoresistible__bu LDALIGQPIS-SGQPVRAPDPVNQPMIRHWAYALSDFNPAYLDPGWAARS
MAB_2054|M.abscessus_ATCC_1997 DASLIGTQLGSTTFP------VDRSKVREFALSLDDHDPIYQDAAAARAA
MAV_3006|M.avium_104 SAARVGTVAASATGE------VNQRDWQRWAAAVGDHNPLYFDPDYARAN
MLBr_02566|M.leprae_Br4923 LRRHFSVKENTVNQPSGLKNILRAIVGALPLLPRTDQLPSRTVTIEELPI
: . * *
Mflv_1511|M.gilvum_PYR-GCK GHPGIVAPPAMIQVWTMGG------LGQGRSDDDPLSKIMELFDEAGYVG
MSMEG_5992|M.smegmatis_MC2_155 GHPGIVAPPAMIQVWTMGG------LGAGRSDDDPLSKIMKLFDDAGYVG
Mvan_5255|M.vanbaalenii_PYR-1 GHPGIVAPPAMIQVWTMGG------LGQGRSDDDPLSKMMKLFDDAGYVG
Mb3572c|M.bovis_AF2122/97 GHPGIVAPPAMIQVWTMMG------LGGVRPKDDPLGPIIKLFDDAGYIG
Rv3542c|M.tuberculosis_H37Rv GHPGIVAPPAMIQVWTMMG------LGGVRPKDDPLGPIIKLFDDAGYIG
MMAR_4247|M.marinum_M RFGGIVSPPVMLQTWTMPAPKLEGIRERGGAPVEIKTNPTAFLDEAGYTS
MUL_0938|M.ulcerans_Agy99 RFGGIVSPPVMLQTWTMPAPKLEGIRERGGAPVEIKTNPTAFLDEAGYTS
TH_4561|M.thermoresistible__bu RYGGIVSPPVMLQSWTMAPPKLQGIHERGGVPVEIRDNPLQFLADAGYTG
MAB_2054|M.abscessus_ATCC_1997 GFGAIPAPPTFVVSS-----------AHWRADDDMFG-----ALGLDLRR
MAV_3006|M.avium_104 GFRDIICPPLFLQYAILGVS-----RLDSLRPDGSSGAVSGSLAFPRAPK
MLBr_02566|M.leprae_Br4923 DHTNVSAYASVTGLRYGNHVP--LTYPFALTFPAMMSLVTGFDFPFAAMG
. : . . .
Mflv_1511|M.gilvum_PYR-GCK VVATNCEQTYHRYLQPGEELTIHADITDVVGP---------KQTALGEGF
MSMEG_5992|M.smegmatis_MC2_155 VVATNCEQTYHRYLQPGEELTIHAEITDVVGP---------KQTALGEGY
Mvan_5255|M.vanbaalenii_PYR-1 VVATNCEQTYHRYLQPGEEVTIAAELTDVIGP---------KQTALGEGF
Mb3572c|M.bovis_AF2122/97 VVATNCEQTYHRYLLPGEQVSISAELGDVVGP---------KQTALGEGW
Rv3542c|M.tuberculosis_H37Rv VVATNCEQTYHRYLLPGEQVSISAELGDVVGP---------KQTALGEGW
MMAR_4247|M.marinum_M TVAVNSEFEIERYPRLGDLISATTVYEEVSEE---------KKTALGTGF
MUL_0938|M.ulcerans_Agy99 TVAVNSEFEIERYPRLGDLISATTVYEEVSEE---------KKTALGTGF
TH_4561|M.thermoresistible__bu TIATNSEFEIERYPRVGDEITAETVFETISDE---------KKTAMGHGF
MAB_2054|M.abscessus_ATCC_1997 VLHGECGWEYFAPVVVGDELTETRRVSNVTSR---------EGKRGGTMT
MAV_3006|M.avium_104 RMAGGESFTFHLPAYHRDRVDMVRTIASIVEK---------QGRS-GRFV
MLBr_02566|M.leprae_Br4923 SVHTENHITQYRPIAVTDVVGVQVHAENLREHRKGLLVDLVTDVSVGNDT
: : : : *
Mflv_1511|M.gilvum_PYR-GCK FINQLITWTVGDGETVAEMNWRIMKFKPRAEDK-PPVPEDLDPDKLMRPA
MSMEG_5992|M.smegmatis_MC2_155 FINQLITWTTEEGEAVAEMNWRIMKFKPREDQK-PAVPDDLDPDKLMRPA
Mvan_5255|M.vanbaalenii_PYR-1 FINQKITWQVGD-EDVAEMMWRIMKFKPAERAKGAQVPEDLDPDKLMRPA
Mb3572c|M.bovis_AF2122/97 FINQHIVWQVGD-EDVAEMNWRILKFKPAGSPS--SVPDDLDPDAMMRPS
Rv3542c|M.tuberculosis_H37Rv FINQHIVWQVGD-EDVAEMNWRILKFKPAGSPS--SVPDDLDPDAMMRPS
MMAR_4247|M.marinum_M FLTWVTTYVDQHGEVLGRQRFRVLRFRPQR--------------------
MUL_0938|M.ulcerans_Agy99 FLTWVTTHVDQHGEALGRQRFRVLRFRPQR--------------------
TH_4561|M.thermoresistible__bu FVTWVTTYRDQHGEVIGRQRFRTLRFKTGS--------------------
MAB_2054|M.abscessus_ATCC_1997 IVTIETDFTNQRDELVVRQTDVLIETGGSTQ-------------------
MAV_3006|M.avium_104 LVTWHTVYRNQHDELLAEASTSMIARP-----------------------
MLBr_02566|M.leprae_Br4923 AWHQVTTFLHLQRTSLSDEPKPPSQKQPKLLPPSAVLQITPRQIRRYAAV
:
Mflv_1511|M.gilvum_PYR-GCK SSKDTRFFWDGVNAHELRIQRRPDGSLQHPPVPAVWADKDAPTDYVVASG
MSMEG_5992|M.smegmatis_MC2_155 SSRDTQFFWDGVNAHELRIQRRPDGTLQHPPVPAVWQDKEQPIDYVVASG
Mvan_5255|M.vanbaalenii_PYR-1 SSRDTQFFWDGVNAHELRIQRRPDGTLQHPPVPAVWQSQDAPIDYAVASG
Mb3572c|M.bovis_AF2122/97 SSRDTAFFWDGVKAHELRIQRLADGSLRHPPVPAVWQDKSVPINYVVSSG
Rv3542c|M.tuberculosis_H37Rv SSRDTAFFWDGVKAHELRIQRLADGSLRHPPVPAVWQDKSVPINYVVSSG
MMAR_4247|M.marinum_M --------------------------------------------------
MUL_0938|M.ulcerans_Agy99 --------------------------------------------------
TH_4561|M.thermoresistible__bu --------------------------------------------------
MAB_2054|M.abscessus_ATCC_1997 --------------------------------------------------
MAV_3006|M.avium_104 --------------------------------------------------
MLBr_02566|M.leprae_Br4923 GGDHNPIHTNPIVAKLFGFPTTIAHGMFSAAAVLANIEARLPDAVHYSAR
Mflv_1511|M.gilvum_PYR-GCK NGTVFSFVVHHAPQVPGRSLPFVIALVELDEGVRMLGELRNVDPADVQIG
MSMEG_5992|M.smegmatis_MC2_155 NGTVFSYVVHHAPKVPGRTLPFVIALVELEEGVRMLGELRNVDPDQVQIG
Mvan_5255|M.vanbaalenii_PYR-1 NGTVYSYVVHHAPKVPGRTLPFVIALVELDEGVRMLGELRGVDPDQVEIG
Mb3572c|M.bovis_AF2122/97 RGTVFSFVVHHAPKVPGRTVPFVIALVELEEGVRMLGELRGADPARVAIG
Rv3542c|M.tuberculosis_H37Rv RGTVFSFVVHHAPKVPGRTVPFVIALVELEEGVRMLGELRGADPARVAIG
MMAR_4247|M.marinum_M --------------------------------------------------
MUL_0938|M.ulcerans_Agy99 --------------------------------------------------
TH_4561|M.thermoresistible__bu --------------------------------------------------
MAB_2054|M.abscessus_ATCC_1997 --------------------------------------------------
MAV_3006|M.avium_104 --------------------------------------------------
MLBr_02566|M.leprae_Br4923 FVKPVVLPATTGLYVDESAGNWDLTLRNIAKGYPHLAGTVQGV-------
Mflv_1511|M.gilvum_PYR-GCK MPVRATYLDFPDSDVSPAWTLYAWEPAR
MSMEG_5992|M.smegmatis_MC2_155 MPVRATFIDFPDSEISPAWTLYAWEPRA
Mvan_5255|M.vanbaalenii_PYR-1 LPVRATYIDFPDSEVSPAWTLYAWEPVR
Mb3572c|M.bovis_AF2122/97 MPVRATYIDFPD------WSLYAWEPDE
Rv3542c|M.tuberculosis_H37Rv MPVRATYIDFPD------WSLYAWEPDE
MMAR_4247|M.marinum_M ----------------------------
MUL_0938|M.ulcerans_Agy99 ----------------------------
TH_4561|M.thermoresistible__bu ----------------------------
MAB_2054|M.abscessus_ATCC_1997 ----------------------------
MAV_3006|M.avium_104 ----------------------------
MLBr_02566|M.leprae_Br4923 ----------------------------