For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
AIIDKETQVRPSFDDEVAATQRYFDDPRFARITRLYTARQVAEQRGTIRTDYTVARDAAAAFYERLRELF AQKKSITTFGPYSPGQAVTMKRMGIEGIYLGGWATSAKGSTTEDPGPDLASYPLSQVPDDAAVLVRALLT ADRNQQYLRLQMSEQQRAATKEYDYRPFIIADADTGHGGDPHVRNLIRRFVEVGVPGYHIEDQRPGTKKC GHQGGKVLVPSDEQIKRLNAARFQLDIMRVPGIIVARTDAEAANLLDSRADERDQPFLLGATNLDIPSYK ACFLAMVRRFYELGVKDLNGHLLYALPEAEYAEATAWLERQGIQGVISDAVNTWRENGQQSIDDLFDQVE SRFVAAWEDDAGLMTYGEAVAEVLEFAASEGEPADMSADEWRAFAARASLYSAKAKAKELGFDPGWDCEL AKTPEGYYQIRGGIPYAIAKSLAAAPFADILWMETKTADLADAKQFADAIHAEFPDQMLAYNLSPSFNWD TTGMTDEQMKQFPEELGKMGFVFNFITYGGHQIDGVAAEEFATSLQQDGMLALARLQRKMRLVESPYRTP QTLVGGPRSDAALAASSGRTATTKAMGEGSTQHQHLVQTEVPKKLLEEWLAMWSENYHLGEKLRVQLRPR RAGSDVLELGIYGDGDEQLANVVVDPIKDRHGRSILQVRDQNTFAEKLRQKRLMTLIHLWLVHRFKADAV IYVTPTEDNLYQTSKMKSHGIFSEVYQEVGEIIVAEVNRPRIAELLQPDRVALRKLITKEG*
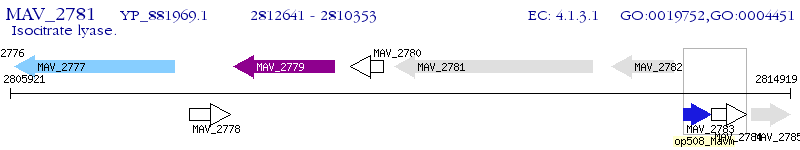
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_2781 | aceA | - | 100% (762) | isocitrate lyase |
| M. avium 104 | MAV_4682 | aceA | 3e-40 | 37.14% (245) | isocitrate lyase |
| M. avium 104 | MAV_0345 | prpB | 9e-12 | 33.71% (175) | methylisocitrate lyase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1950 | aceA | 0.0 | 86.27% (765) | isocitrate lyase |
| M. gilvum PYR-GCK | Mflv_3260 | - | 0.0 | 80.03% (761) | isocitrate lyase |
| M. tuberculosis H37Rv | Rv1916 | aceAb | 0.0 | 87.15% (397) | isocitrate lyase |
| M. leprae Br4923 | MLBr_01985 | aceA | 0.0 | 87.40% (762) | isocitrate lyase |
| M. abscessus ATCC 19977 | MAB_4095c | - | 3e-37 | 35.60% (250) | isocitrate lyase (AceA) |
| M. marinum M | MMAR_2821 | aceAb | 0.0 | 88.03% (760) | isocitrate lyase AceAb |
| M. smegmatis MC2 155 | MSMEG_3706 | aceA | 0.0 | 81.62% (751) | isocitrate lyase |
| M. thermoresistible (build 8) | TH_0417 | aceAb | 0.0 | 80.26% (765) | PROBABLE ISOCITRATE LYASE aceAb |
| M. ulcerans Agy99 | MUL_2943 | aceAb | 0.0 | 87.37% (760) | isocitrate lyase AceAb |
| M. vanbaalenii PYR-1 | Mvan_2977 | - | 0.0 | 81.95% (748) | isocitrate lyase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3260|M.gilvum_PYR-GCK ------------------------------------MAITEADLQS----
Mvan_2977|M.vanbaalenii_PYR-1 ------------------------------------MAIIEADAFQPHAT
MSMEG_3706|M.smegmatis_MC2_155 ------------------------------------MATIEANAHNLSSS
Mb1950|M.bovis_AF2122/97 ------------------------------------MAIAETDTEVHTPF
Rv1916|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_2821|M.marinum_M ------------------------------------MAIAETDTEVRTP-
MUL_2943|M.ulcerans_Agy99 ------------------------------------MAIVETDTEIRTP-
MLBr_01985|M.leprae_Br4923 MAGIPATVVQRCNSTNLDDRLHYSPLGDRAKEERNPMAIMDTNTEVHTL-
MAV_2781|M.avium_104 ------------------------------------MAIIDKETQVRPS-
TH_0417|M.thermoresistible__bu ------------------------------------MAISEAERETRTS-
MAB_4095c|M.abscessus_ATCC_199 ------------------------------------MSNVGK--------
Mflv_3260|M.gilvum_PYR-GCK QTPFERDVVETQRYFDS-PRFEGIIRLYTARQVAEQRGTIPADYPVAREA
Mvan_2977|M.vanbaalenii_PYR-1 GEPFERDVAETQRYFDS-PRFDGITRLYTARQVAEQRGTIPADYPTAREA
MSMEG_3706|M.smegmatis_MC2_155 AKDFDSEVAATQAYFDS-PRFEGITRLYSARQVVEQRGTIATDHTVAREA
Mb1950|M.bovis_AF2122/97 EQDFEKDVAATQRYFDS-SRFAGIIRLYTARQVVEQRGTIPVDHIVAREA
Rv1916|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_2821|M.marinum_M ---FEQEVADTQRYFDS-SRFAGIVRLYTARQVVEQRGTIPNDYTVARTA
MUL_2943|M.ulcerans_Agy99 ---FEQEVADTQRYFDS-SRFAGIVRLYTARQVVEQRGTIPNDYTVARTA
MLBr_01985|M.leprae_Br4923 ---FTQEVAATQQYFDD-PRFAGIIRLYTARQVVEQRGTIPTDYTVARDA
MAV_2781|M.avium_104 ---FDDEVAATQRYFDD-PRFARITRLYTARQVAEQRGTIRTDYTVARDA
TH_0417|M.thermoresistible__bu ---FEEDVAETQRYFDS-PRFDGIIRLYTARQVVEQRGTIPVDYTIAREA
MAB_4095c|M.abscessus_ATCC_199 ----PRTAAEIQQDWDTNPRWKGITRDYTAAQVEELQGSVVEENTLARRG
Mflv_3260|M.gilvum_PYR-GCK ATAFFPHLRELFGQGRSITTFGPYSPGQAVVMKRMGIEGIYLGGWATSAK
Mvan_2977|M.vanbaalenii_PYR-1 ATAFYPHLRELFAKGKSITTFGPYSPGQAVVMKRMGIEGVYLGGWATSAK
MSMEG_3706|M.smegmatis_MC2_155 AEAFHPYLRELFAQKKSITTFGPYSPGQAVVMKRMGIGGIYLGGWATSAK
Mb1950|M.bovis_AF2122/97 AGAFYERLRELFAARKSITTFGPYSPGQAVSMKRMGIEAIYLGGWATSAK
Rv1916|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_2821|M.marinum_M AGAFYQRLRELYAEKKSVTTFGPYSPGQAVSMKRMGIEAIYLGGWATSAK
MUL_2943|M.ulcerans_Agy99 AGVFYQRLRELYAEKKSVTTFGPYSPGQAVSMKRMGIEAIYLGGWATSAK
MLBr_01985|M.leprae_Br4923 ATAFYARLRELFAAGKSVTTFGPYSPGQAVSLKRMGIEAIYLGGWATSAK
MAV_2781|M.avium_104 AAAFYERLRELFAQKKSITTFGPYSPGQAVTMKRMGIEGIYLGGWATSAK
TH_0417|M.thermoresistible__bu ATAFYARLRELFAERKCITTFGPYSPGQAVTMKRMGIEGIYLGGWATSAK
MAB_4095c|M.abscessus_ATCC_199 AEILWDAVSK--DDDSYINALGALTGNMAVQQVRAGLKAIYLSGWQVAGD
Mflv_3260|M.gilvum_PYR-GCK GSISEDPGPDLASYPLSQVPEEAAGIVRALLTADRNQQYLRLQMTPEQQA
Mvan_2977|M.vanbaalenii_PYR-1 GSISEDPGPDLASYPLSQVPEEAASIVRALLTADRNQHYLRLQMDPEQRA
MSMEG_3706|M.smegmatis_MC2_155 GSISEDPGPDLASYPLSQVPDEAAGLVRALLTADRNQHYLRLQMTPEQRE
Mb1950|M.bovis_AF2122/97 GSSTEDPGPDLASYPLSQVPDDAAVLVRALLTADRNQHYLRLQMSERQRA
Rv1916|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_2821|M.marinum_M GSTTEDPGPDLASYPLSQVPDDAAVLVRALLTADRNQEYLRLQMTDQQRA
MUL_2943|M.ulcerans_Agy99 GSTTEDPGPDLASYPLSQVPDDAAVLVRALLTADRNQEYLRLQMTDQQRA
MLBr_01985|M.leprae_Br4923 GSITEDPGPDLASYPLSQVPDDAAVLVRALLAADRNQQYLRLHMTEQQRA
MAV_2781|M.avium_104 GSTTEDPGPDLASYPLSQVPDDAAVLVRALLTADRNQQYLRLQMSEQQRA
TH_0417|M.thermoresistible__bu GSADEDPGPDLASYPLSQVPNEAAGLVRALLTADRNQEYLRQRMSPEQRA
MAB_4095c|M.abscessus_ATCC_199 ANLSGHTYPDQSLYPANSVPAVVRRINNALLRADEIAR---------VEG
Mflv_3260|M.gilvum_PYR-GCK TTPAYDYRPFIIADADTGHGGDPHVRNLIRRFVENGVPGYHIEDQRPGTK
Mvan_2977|M.vanbaalenii_PYR-1 AAPEYDYRPFIIADADTGHGGDPHVRNLIRRFVEAGVPGYHIEDQRPGTK
MSMEG_3706|M.smegmatis_MC2_155 AQPPVDYRPFIIADADTGHGGDPHVRNLIRRFVESGVPGYHIEDQRPGTK
Mb1950|M.bovis_AF2122/97 ATPAYDFRPFIIADADTGHGGDPHVRNLIRRFVEVGVPGYHIEDQRPGTK
Rv1916|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_2821|M.marinum_M TTTAYDFRPFIIADADTGHGGDPHVRNLIRRFVEVGVPGYHIEDQRPGTK
MUL_2943|M.ulcerans_Agy99 TTTAYDFRPFIIADADTGHGGDPHVRNLIRRFVEVGVPGYHIEDQRPGTK
MLBr_01985|M.leprae_Br4923 ATPAYDYRPFIIADADTGHGGDSHVRNLIRRFVEIGVPGYHIEDQRPGTK
MAV_2781|M.avium_104 ATKEYDYRPFIIADADTGHGGDPHVRNLIRRFVEVGVPGYHIEDQRPGTK
TH_0417|M.thermoresistible__bu ATPVYDYRPFIIADADTGHGGDAHVRNLIRRFVEAGVAGYHIEDQRPGTK
MAB_4095c|M.abscessus_ATCC_199 DTSVDNWLVPIVADGEAGFGGALNVYELQKAMIAAGAAGTHWEDQLASEK
Mflv_3260|M.gilvum_PYR-GCK KCGHQGGKVLVPSDEQIKRLNTARFQLDIMRVPGIIVARTDAEAANLIDS
Mvan_2977|M.vanbaalenii_PYR-1 KCGHQGGKVLVPSDEQIKRLNTARFQLDIMRVPGIIVARTDAEAANLIDS
MSMEG_3706|M.smegmatis_MC2_155 KCGHQGGKVLVPSDEQIKRLNTARFQLDIMRVPGIIVARTDAEAANLIDS
Mb1950|M.bovis_AF2122/97 KCGHQGGKVLVPSDEQIKRLNAARFQLDIMRVPGIIVARTDAEAANLIDS
Rv1916|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_2821|M.marinum_M KCGHQGGKVLVPSDEQIKRLNAARFQLDVMRVPGIIVARTDAEAANLIDS
MUL_2943|M.ulcerans_Agy99 KCGHQGGKVLVPSDEQIKRLNAARFQLDVMRVPGIIVARTDAEAANLIDS
MLBr_01985|M.leprae_Br4923 KCGHQGGKVLVPSDEQIKRLNAARFQLDIMRVPGIIVARTDAEAANLIDS
MAV_2781|M.avium_104 KCGHQGGKVLVPSDEQIKRLNAARFQLDIMRVPGIIVARTDAEAANLLDS
TH_0417|M.thermoresistible__bu KCGHQGGKVLVPSDEQIKRLNTARFQLDIMRVPGIIVARTDAEAANLIDS
MAB_4095c|M.abscessus_ATCC_199 KCGHLGGKVLIPTQQHIRTLTSARLAADVANTPTVVIARTDAEAATLITS
Mflv_3260|M.gilvum_PYR-GCK RADERDQPFLLGATNLKIPSYKSCYLAMMRRFYDKGVTELNGHLLYALPD
Mvan_2977|M.vanbaalenii_PYR-1 RADERDQPFLLGATNLKIPSYKSCFLAMVRRFYEKGMTELNGHLLYALPE
MSMEG_3706|M.smegmatis_MC2_155 RADERDQPFLLGVTNLKIPSYKSCFLAMMRRFYDQGLTEINGHLLYDLPE
Mb1950|M.bovis_AF2122/97 RADERDQPFLLGATKLDVPSYKSCFLAMVRRFYELGVKELNGHLLYALGD
Rv1916|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_2821|M.marinum_M RADERDQPFLLGATKLDIPSYKSCFLAMLRRFYELGVKELNGHLLYALAD
MUL_2943|M.ulcerans_Agy99 RADERDQPFLLGATKLDIPSYKSCFLAMLRRFYELGVKELNGHLLYALAD
MLBr_01985|M.leprae_Br4923 RADERDQPFLLGATNLKIPSYKACFLALVRCFYELGVKELHGHLLYALGD
MAV_2781|M.avium_104 RADERDQPFLLGATNLDIPSYKACFLAMVRRFYELGVKDLNGHLLYALPE
TH_0417|M.thermoresistible__bu RADERDQPFLLGATNLAIPSYKSCYLAMLRSFYDSGVTELNGHLLYWLAE
MAB_4095c|M.abscessus_ATCC_199 DVDERDKPFVTG--------------------------------------
Mflv_3260|M.gilvum_PYR-GCK GEYATAEAWLQRQGVLDLVDQTAAAWQDGTEASLDTAFDKVESRFVEAWQ
Mvan_2977|M.vanbaalenii_PYR-1 GEYATATAWLDRQGVLNLIDETASAWQAGTESSVDAAFDRVESRFVDAWQ
MSMEG_3706|M.smegmatis_MC2_155 GQYEAAEAWLTGAGIMDEIAAAVESWTSGAASSVDALYDAVESKFVDAWQ
Mb1950|M.bovis_AF2122/97 SEYAAAGGWLERQGIFGLVSDAVNAWREDGQQSIDGIFDQVESRFVAAWE
Rv1916|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_2821|M.marinum_M AEYAAANGWLERSGIMSLISDAVNSWREDGMQTIDDLFDQVESRFVAAWE
MUL_2943|M.ulcerans_Agy99 AEYAAANGWLERSGIMGLISDAVNSWREDGMQTTDDLFDQVESRFVAAWE
MLBr_01985|M.leprae_Br4923 GEYAAASAWLDRQGILAQVSGTVNAWQENGKQSIDDLFEQVEYRLLAAWE
MAV_2781|M.avium_104 AEYAEATAWLERQGIQGVISDAVNTWRENGQQSIDDLFDQVESRFVAAWE
TH_0417|M.thermoresistible__bu GEYDRARAWLDRNGLLDRISEAVGKWRPGEDPANDELFDEVESAFVEIWE
MAB_4095c|M.abscessus_ATCC_199 --------------------------------------------------
Mflv_3260|M.gilvum_PYR-GCK DDAGLNTYGEAVAELLEFREREGEPSPMSVAQWREFAERAPLYTAREKAK
Mvan_2977|M.vanbaalenii_PYR-1 DDAGLNTYGEAVAELLEFREREGEPSSMSVADWRMFAERAPLYTAREKAK
MSMEG_3706|M.smegmatis_MC2_155 DDAGLNTYGEAVAEVIEFREREGESVAMSAAEWREFATRAPLYTATNKAR
Mb1950|M.bovis_AF2122/97 DDAGLMTYGEAVADVLEFGQSEGEPIGMAPEEWRAFAARASLHAARAKAK
Rv1916|M.tuberculosis_H37Rv -----MTYGEAVADVLEFGQSEGEPIGMAPEEWRAFAARASLHAARAKAK
MMAR_2821|M.marinum_M DDAGLMTYAEAVAEVLAFGQSEGDPVAMSPEEWRTFAARASLYAARQKAK
MUL_2943|M.ulcerans_Agy99 DDAGLMTYAEAVAEVLAFGQSEGDPVPMSPEEWRTFAARASLYAARQKAK
MLBr_01985|M.leprae_Br4923 KDAGLMTYGEAVEEMLQFGESEGELIGMSPEEWRRFVGRASLYAAREKAK
MAV_2781|M.avium_104 DDAGLMTYGEAVAEVLEFAASEGEPADMSADEWRAFAARASLYSAKAKAK
TH_0417|M.thermoresistible__bu EDAGLKTYGEAIADRMEFAEADGEPFDMTAEEWRRFAERKSFFEMSQKAA
MAB_4095c|M.abscessus_ATCC_199 --------------------------------------------------
Mflv_3260|M.gilvum_PYR-GCK ELGADVAWDCERVKTPEGYYQVRGGIPYAIAKSLAAAPFADILWMETKTA
Mvan_2977|M.vanbaalenii_PYR-1 ELGADVAWDCERVKTPEGYYQVRGGIPYAIAKSLAAAPFADILWMETKTA
MSMEG_3706|M.smegmatis_MC2_155 ELGVDVAWDCEPAKTPEGYYQVRGGIPYAIAKSLAAAPFADILWMETKTA
Mb1950|M.bovis_AF2122/97 ELGADPPWDCELAKTPEGYYQIRGGIPYAIAKSLAAAPFADILWMETKTA
Rv1916|M.tuberculosis_H37Rv ELGADPPWDCELAKTPEGYYQIRGGIPYAIAKSLAAAPFADILWMETKTA
MMAR_2821|M.marinum_M ELGVDPGWDCELAKTPEGYYQIRGGIPYAIAKSLAAAPFADILWMETKTA
MUL_2943|M.ulcerans_Agy99 EMGVDPGWDCELAKTPEGYYQIRGGIPYAIAKSLAAAPFADILWMETKTA
MLBr_01985|M.leprae_Br4923 ELGVDPGWDCELAKTPEGYYQIRGGIQYAIAKSLAAAPFADILWMETKTA
MAV_2781|M.avium_104 ELGFDPGWDCELAKTPEGYYQIRGGIPYAIAKSLAAAPFADILWMETKTA
TH_0417|M.thermoresistible__bu ELGIDVPWDCELAKTPEGYYQLRGGIPYAIAKSLAAAPFADLLWMETKTA
MAB_4095c|M.abscessus_ATCC_199 ------------ERTSEGFYNVQKGIEPCIARAKAYAPYADLIWMETGVP
:*.**:*::: ** .**:: * **:**::**** ..
Mflv_3260|M.gilvum_PYR-GCK DLADAKQFADAIHAVYPDKMLAYNLSPSFNWDTTGMTDEEMRAFPEEIGK
Mvan_2977|M.vanbaalenii_PYR-1 DLADAKQFADAIHAVYPDKMLAYNLSPSFNWDTTGMSDEEMRTFPEEIGK
MSMEG_3706|M.smegmatis_MC2_155 DLADAKQFADAIHAVYPDQMLAYNLSPSFNWDTTGMTDDEMRAFPEELGK
Mb1950|M.bovis_AF2122/97 DLADARQFAEAIHAEFPDQMLAYNLSPSFNWDTTGMTDEEMRRFPEELGK
Rv1916|M.tuberculosis_H37Rv DLADARQFAEAIHAEFPDQMLAYNLSPSFNWDTTGMTDEEMRRFPEELGK
MMAR_2821|M.marinum_M DLADARQFAEAIHAEFPDQMLAYNLSPSFNWDTTGMTDEEMKRFPEELGK
MUL_2943|M.ulcerans_Agy99 DLADARQFAEAIHAEFPDQMLAYNLSPSFNWDTTGMTDEEMKRFPEELGK
MLBr_01985|M.leprae_Br4923 DLADARQFAEAIHAEFPEQMLAYNLSPSFNWDTTGMSDEEMKRFPEELGK
MAV_2781|M.avium_104 DLADAKQFADAIHAEFPDQMLAYNLSPSFNWDTTGMTDEQMKQFPEELGK
TH_0417|M.thermoresistible__bu DLDDARQFAEAIHAQFPDKMLAYNLSPSFNWDTTGMTDEEMRAFPAELGK
MAB_4095c|M.abscessus_ATCC_199 DLEVARKFAEAVKADFPDQLLSYNCSPSFNWKQH-LDDSTIAKFQKELGA
** *::**:*::* :*:::*:** ******. : *. : * *:*
Mflv_3260|M.gilvum_PYR-GCK MGFVFNFITYGGHQVDGVASEEFATSLRQEGMLALARLQRKMRLVESPYR
Mvan_2977|M.vanbaalenii_PYR-1 MGFVFNFITYGGHQVDGVASEEFATSLRQEGMLALARLQRKMRLVESPYR
MSMEG_3706|M.smegmatis_MC2_155 MGFVFNFITYGGHQVDGVACEEFATSLRQEGMLALARLQRKMRLVESPYR
Mb1950|M.bovis_AF2122/97 MGFVFNFITYGGHQIDGVAAEEFATALRQDGMLALARLQRKMRLVESPYR
Rv1916|M.tuberculosis_H37Rv MGFVFNFITYGGHQIDGVAAEEFATALRQDGMLALARLQRKMRLVESPYR
MMAR_2821|M.marinum_M MGFVFNFITYGGHQIDGVASEEFATALRQDGMLALARLQRKMRLVESPYR
MUL_2943|M.ulcerans_Agy99 MGFVFNFITYGGHQIDGVASEEFATALRQDGMLALARLQRKMRLVESPYR
MLBr_01985|M.leprae_Br4923 MGFVFNFITYGGHQIDGVAAEEFATALRQDGMLALARLQRKMRLVESPYR
MAV_2781|M.avium_104 MGFVFNFITYGGHQIDGVAAEEFATSLQQDGMLALARLQRKMRLVESPYR
TH_0417|M.thermoresistible__bu MGFVFNFITYGGHQIDGVAAEEFATALRQDGMLALARIQRKLRLIESPYR
MAB_4095c|M.abscessus_ATCC_199 MGFKFQFITLAGFHALNYSMFDLAYGYAREGMTAYVDLQ-----------
*** *:*** .*.: . : ::* . ::** * . :*
Mflv_3260|M.gilvum_PYR-GCK TPQTLVGGPRSDAALAASSGRTATTKSMGKGSTQHQHLLQTEVPKKLLEE
Mvan_2977|M.vanbaalenii_PYR-1 TPQTLVGGPRSDAALAASSGRTATTKSMGKGSTQHQHLVQTEVPKKLLEE
MSMEG_3706|M.smegmatis_MC2_155 TPQTLVGGPRSDAALAASSGRTATTKAMGKGSTQHQHLVQTEVPKKLLEE
Mb1950|M.bovis_AF2122/97 TPQTLVGGPRSDAALAASSGRTATTKAMGKGSTQHQHLVQTEVPRKLLEE
Rv1916|M.tuberculosis_H37Rv TPQTLVGGPRSDAALAASSGRTATTKAMGKGSTQHQHLVQTEVPRKLLEE
MMAR_2821|M.marinum_M TPQTLVGGPRSDAALQASSGRTATTKAMGKGSTQHQHLVQTEVPKKLLEE
MUL_2943|M.ulcerans_Agy99 TPQTLVAGPRSDAALQASSGRTATTKAMGKGSTQHQHLVQTEVPKKLLEE
MLBr_01985|M.leprae_Br4923 TPQTLVGGPRSDAALAASSGRTATTKAMGKGSTQHQHLVQTEVPKKLLEE
MAV_2781|M.avium_104 TPQTLVGGPRSDAALAASSGRTATTKAMGEGSTQHQHLVQTEVPKKLLEE
TH_0417|M.thermoresistible__bu TPQTLVGGPRSDAALTASSGRTATTKAMGEGSTQHQHLVQTEVPKKLLEE
MAB_4095c|M.abscessus_ATCC_199 -------------------EREFASEARGYTATKHQREVG----------
* :::: * :*:**: :
Mflv_3260|M.gilvum_PYR-GCK WLALWSEHNKLPERLHVQLRPRRAGSDVLDLQILG----DSEDPLANVVV
Mvan_2977|M.vanbaalenii_PYR-1 WLGLWSEHYQLPERLHVQLRPRRAGSDVLDLQILG----DSEEPLANVVV
MSMEG_3706|M.smegmatis_MC2_155 WLAMWSEHYGLPEKLHVQLRPRRAGSDVLDLQILGEG-GDGTDPLANVVV
Mb1950|M.bovis_AF2122/97 WLAMWSGHYQLKDKLRVQLRPQRAGSEVLELGIHGE----SDDKLANVIF
Rv1916|M.tuberculosis_H37Rv WLAMWSGHYQLKDKLRVQLRPQRAGSEVLELGIHGE----SDDKLANVIF
MMAR_2821|M.marinum_M WLALWSDHYQLGEKLRVQLRPRRAGSDVLELGIYGN----DDEQLANVVF
MUL_2943|M.ulcerans_Agy99 RLALWSDHYQLGEKLRVQLRPRRAGSDVLELGIYGN----DDEQLANVVF
MLBr_01985|M.leprae_Br4923 WLALWSEHYQLGEKLRVQLRPRRAGSDVLELGIYGN----GDEQLANVIV
MAV_2781|M.avium_104 WLAMWSENYHLGEKLRVQLRPRRAGSDVLELGIYGD----GDEQLANVVV
TH_0417|M.thermoresistible__bu WLGMWSEHYQLGEKLRVQLRPRRAGGEVLELGIFAERPGGEDEKVANVLV
MAB_4095c|M.abscessus_ATCC_199 --------------------------------------------------
Mflv_3260|M.gilvum_PYR-GCK DPIKDRHGRSILTVRDQNTFAEHLRQKRLMDVIHLWLIHRFKAEIVYYVT
Mvan_2977|M.vanbaalenii_PYR-1 DPIKDRHGRSILTVRDQNTFAEQMRQKRLMDLIHLWLIHRFKAEIVYYVT
MSMEG_3706|M.smegmatis_MC2_155 DPIKDRHGRSILTVRDQNTFAEQMRKKRLMDLIHLWLIHRFKAEIVYYVT
Mb1950|M.bovis_AF2122/97 QPIQDRRGRTILLVRDQNTFGAELRQKRLMTLIHLWLVHRFKAQAVHYVT
Rv1916|M.tuberculosis_H37Rv QPIQDRRGRTILLVRDQNTFGAELRQKRLMTLIHLWLVHRFKAQAVHYVT
MMAR_2821|M.marinum_M DPIKDRHGRSILTVRDQNTFAHKLRQKRLMTLIHLWLVHRFKADAVYYVT
MUL_2943|M.ulcerans_Agy99 DPIKDRHGRSILTVRDQNTFAHKLRQKRLMTLIHLWLVHRFKADAVYYVT
MLBr_01985|M.leprae_Br4923 DPIKDRRGRSILQVRDQNTFAEKLRQKRLMTLIHFWLVHRFKADTVIYVT
MAV_2781|M.avium_104 DPIKDRHGRSILQVRDQNTFAEKLRQKRLMTLIHLWLVHRFKADAVIYVT
TH_0417|M.thermoresistible__bu DPIRDRHGRSILTVRDQNTFAEALRKKRLMTLIHLWLVHRFKAEAVYYVS
MAB_4095c|M.abscessus_ATCC_199 ----AGYFDKIATTVDPNTSTAALKG------------------------
.* . * ** ::
Mflv_3260|M.gilvum_PYR-GCK PTEDNIYQTEKMKSHGIFSDVYQEVGEIIVADVNQARIDELLAPDREALT
Mvan_2977|M.vanbaalenii_PYR-1 PTEDNIYQTEKMKSHGIFSDVYQEVGEIIVADVNQARIDELLAPDRKALI
MSMEG_3706|M.smegmatis_MC2_155 PTEDNIYQTEKMKAHGIFSDVYQEVGEIIVADVNKPRIEELLASDREALG
Mb1950|M.bovis_AF2122/97 PTDDNLYQTSKMKSHGIFTEVNQEVGEIIVAEVNHPRIAELLTPDRVALR
Rv1916|M.tuberculosis_H37Rv PTDDNLYQTSKMKSHGIFTEVNQEVGEIIVAEVNHPRIAELLTPDRVALR
MMAR_2821|M.marinum_M PTEDNQYQTSKMKSHGIFSEVNQDVGEIIVAEVNRPRIEELLTADRVALR
MUL_2943|M.ulcerans_Agy99 PTEDNQYQTSKMKSHGIFSEVNQDVGEIIVAEVNRPRIEELLTADRVALR
MLBr_01985|M.leprae_Br4923 PTEDNLYQTSKMKSHGIFSEVYQEVGEIIVAEVNHPRLVELLTPDRVALR
MAV_2781|M.avium_104 PTEDNLYQTSKMKSHGIFSEVYQEVGEIIVAEVNRPRIAELLQPDRVALR
TH_0417|M.thermoresistible__bu PTEDNLYQTSKMKSHGIFSEVHQDVGEIIVADVNRERIEELLAPDREALR
MAB_4095c|M.abscessus_ATCC_199 STEEGQFH------------------------------------------
.*::. ::
Mflv_3260|M.gilvum_PYR-GCK RLITKQD
Mvan_2977|M.vanbaalenii_PYR-1 RLINKQD
MSMEG_3706|M.smegmatis_MC2_155 RLIRKED
Mb1950|M.bovis_AF2122/97 KLITKEA
Rv1916|M.tuberculosis_H37Rv KLITKEA
MMAR_2821|M.marinum_M QLITKGD
MUL_2943|M.ulcerans_Agy99 QLITKGD
MLBr_01985|M.leprae_Br4923 RLITKEG
MAV_2781|M.avium_104 KLITKEG
TH_0417|M.thermoresistible__bu RLIRKED
MAB_4095c|M.abscessus_ATCC_199 -------