For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MQTADWRPRLELLLAEFRRRQADVARSGVEPDRIEVARAWHAELVDHQLAAPGWPREVGGLDLSLADQLD YYRMTTDAGVPPHPCPLSFILAPTLIAHGTQQQKDRFLIPLLRADEFWCQGFSEPGAGSDLSSLSTRAVR DGDVYRVTGQKVWTTMADRADWMFALVRTGSGGKPSDGITYLLIPMKSPGITVRPLRDISGAAHFAEVFL DDVAVPVENVVGDEGAGWSIMRTSLGHERATAFLADEFKYRRTADRVIDLVVAQRLDDDPLVRQDVARLE SGVRTIAANSARALAAVLRGEDPGGMASVNRLVKSEFEQHMHALALRAAGPYAALGSRAPDAVDKGRWTF GYLMSRATTIGAGTAEIQRNTIAESVLGLPSHRGEGTREAAVTPGAPLAVPEEDERELREVLAATLQAKV DEAALLDRKRPVDAAEPEVWSALVEFGLPGLAVDETLGGAGARPRLLYTAVEEAAKALAPAPLVPTVIGL DVALHCGAKALVQRITGGATAAFAVPVNDSGWVTSGPELPEWDGTGLSGVVPIVAGAPHAEVLVALARVA AGVGEVLVAVDASADGVEVTAQQPLDLTGTVGAVTFSGAAGEVLADGTDVPRMLAAARRRAALAVAADSV GVASRALAMAVQWARERHQFGRPIGSFQAVSHRCADMLVALEGARSQILEAAESDLEESGYLADLAAAAA LDAGAAATEGALQIHGGIGFTWEHPIHLLLRRARANSVLVGRADALRDRAAGELLAPYRQQRSE
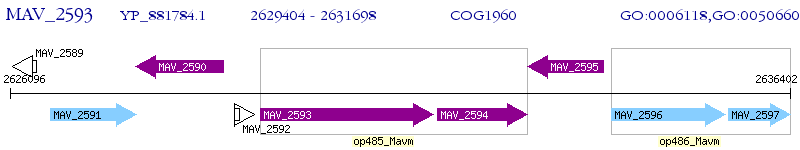
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_2593 | - | - | 100% (764) | acyl-CoA dehydrogenase |
| M. avium 104 | MAV_1877 | - | 0.0 | 76.92% (754) | acyl-CoA dehydrogenase |
| M. avium 104 | MAV_0601 | - | 6e-63 | 38.14% (354) | putative acyl-CoA dehydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3590c | fadE30 | 2e-63 | 38.94% (357) | acyl-CoA dehydrogenase FADE30 |
| M. gilvum PYR-GCK | Mflv_1481 | - | 5e-64 | 37.57% (354) | acyl-CoA dehydrogenase domain-containing protein |
| M. tuberculosis H37Rv | Rv3560c | fadE30 | 2e-63 | 38.94% (357) | acyl-CoA dehydrogenase FADE30 |
| M. leprae Br4923 | MLBr_00737 | fadE25 | 1e-15 | 23.42% (363) | putative acyl-CoA dehydrogenase |
| M. abscessus ATCC 19977 | MAB_0597 | - | 2e-61 | 35.85% (371) | acyl-CoA dehydrogenase FadE |
| M. marinum M | MMAR_5049 | fadE30 | 2e-60 | 37.75% (355) | acyl-CoA dehydrogenase FadE30 |
| M. smegmatis MC2 155 | MSMEG_6012 | - | 5e-58 | 35.16% (364) | putative acyl-CoA dehydrogenase |
| M. thermoresistible (build 8) | TH_4120 | - | 4e-67 | 41.42% (367) | PUTATIVE putative acyl-CoA dehydrogenase domain protein |
| M. ulcerans Agy99 | MUL_4125 | fadE30 | 6e-60 | 37.46% (355) | acyl-CoA dehydrogenase FadE30 |
| M. vanbaalenii PYR-1 | Mvan_5285 | - | 9e-62 | 37.36% (356) | acyl-CoA dehydrogenase domain-containing protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1481|M.gilvum_PYR-GCK --------------MIEV--QKFRAEVRDWLAENLVGDFAALKGLGGPGR
Mvan_5285|M.vanbaalenii_PYR-1 --------------MIEV--QEFRAEVRDWLAENLVGEYAALKGLGGPGR
MSMEG_6012|M.smegmatis_MC2_155 --------------MIEV--QEFRAEVRQWLADNLVGDFAALKGLGGPGR
Mb3590c|M.bovis_AF2122/97 --------------MQDV--EEFRAQVRGWLADNLAGEFAALKGLGGPGR
Rv3560c|M.tuberculosis_H37Rv --------------MQDV--EEFRAQVRGWLADNLAGEFAALKGLGGPGR
MMAR_5049|M.marinum_M --------------MQDV--EEFRAEVRGWLAANLVGEFATLKGLGGPGR
MUL_4125|M.ulcerans_Agy99 --------------MQDV--EEFRAEVRGWLAANLVGEFATLKGLGGPGR
MAB_0597|M.abscessus_ATCC_1997 MVESQVSPAPAGEPMSEMSDDQVRTEIREWLAENLVGEFAELKGLGGPGS
TH_4120|M.thermoresistible__bu -----VNDRDYGVALTDAD-RAFRDEVREFLASSLTPDLLSRD-----GG
MAV_2593|M.avium_104 ---------------------MQTADWRPRLELLLAEFRRRQADVARSGV
MLBr_00737|M.leprae_Br4923 ----------MVGWSGNPLFDLFKLPEEHNELRATIRALAEKEIAPHAAD
. .
Mflv_1481|M.gilvum_PYR-GCK EHEAFEERLAWNQHLAAAGLTCLGWPEEHGGRGLSVAHRVAFYEEYAKAN
Mvan_5285|M.vanbaalenii_PYR-1 EHEAFEERLAWNRHLAAAGLTCLGWPEEHGGRGLSVAHRVAFYEEYAKAN
MSMEG_6012|M.smegmatis_MC2_155 EHEAFEERRAWNQHLAAAGLTCLGWPVEHGGRGLSVAHRVAFYEEYARAN
Mb3590c|M.bovis_AF2122/97 EHEAFEERRAWNQRLAAAGLTCLGWPEEHGGRGLSTAHRVAFYEEYARAD
Rv3560c|M.tuberculosis_H37Rv EHEAFEERRAWNQRLAAAGLTCLGWPEEHGGRGLSTAHRVAFYEEYARAD
MMAR_5049|M.marinum_M EHEAFEERRAWNQHLAAAGLTCLGWPVEHGGRGLSTAHRVAFYEEYARAD
MUL_4125|M.ulcerans_Agy99 EHEAFEERRAWNQHLAAAGLTCLGWPVEHGGRGLSTAHRVAFYEEYARAD
MAB_0597|M.abscessus_ATCC_1997 EHEAFEERLAWNRHLAAAGWTCLGWPVEHGGRGATVSQRVIFYEEYARAE
TH_4120|M.thermoresistible__bu ETARLDRLRRWQGLLYDAGLAAISWPTEYGGRAATPTQQLIFNTEMAAAR
MAV_2593|M.avium_104 EPDRIEVARAWHAELVDHQLAAPGWPREVGGLDLSLADQLDYYRMTTDAG
MLBr_00737|M.leprae_Br4923 VDQRARFPEEALAALNASGFNAIHVPEEYGGQGADSVAACIVIEEVARVD
* . * * ** : .
Mflv_1481|M.gilvum_PYR-GCK APDKVNHLGEELLGPTLIAYGTPEQQQRFLPRILDVTELWSQGYSEPGAG
Mvan_5285|M.vanbaalenii_PYR-1 APDKVNHLGEELLGPTLIAYGTPEQQQRFLPKILDVTELWSQGYSEPGAG
MSMEG_6012|M.smegmatis_MC2_155 APDKVNHFGEELLGPTLIEYGTPEQQKRFLPKILDVTELWCQGYSEPNAG
Mb3590c|M.bovis_AF2122/97 APDKVNHFGEELLGPTLIAFGTPQQQRRFLPRIRDVTELWCQGYSEPGAG
Rv3560c|M.tuberculosis_H37Rv APDKVNHFGEELLGPTLIAFGTPQQQRRFLPRIRDVTELWCQGYSEPGAG
MMAR_5049|M.marinum_M APDKVNHFGEELLGPTLIAFGTPEQQQRFLPRILDVTELWCQGYSEPGAG
MUL_4125|M.ulcerans_Agy99 APDKVNHFGEELLGPTLIAFGTPEQQQRFLPRILDVTELWCQGYSEPGAG
MAB_0597|M.abscessus_ATCC_1997 APHKVNHLGEELLGPTLIAYGTPEQQQRFLPAIRDVTELWCQGYSEPGAG
TH_4120|M.thermoresistible__bu APEPINRSAINQLGPTIIQWGTDEQRAYYLPRILSAEDVWCQGFSEPEAG
MAV_2593|M.avium_104 VPPHP-CPLSFILAPTLIAHGTQQQKDRFLIPLLRADEFWCQGFSEPGAG
MLBr_00737|M.leprae_Br4923 ASASLIPAVNKLGTMGLILRGSEELKKQVLPSLAAEGAMASYALSEREAG
.. :* *: : : * : . . . ** **
Mflv_1481|M.gilvum_PYR-GCK SDLANVSTTATLD----------GDEWVINGQKVWTSLAHWAQWCFVVAR
Mvan_5285|M.vanbaalenii_PYR-1 SDLANVSTTAVLDE--------EGRQWHINGQKVWTSLAHWAQWCFVVAR
MSMEG_6012|M.smegmatis_MC2_155 SDLANVSTTAELVGDAEASGATGNQYWVINGQKVWTSLAHWAQWCFVVAR
Mb3590c|M.bovis_AF2122/97 SDLASVATTAELD----------GDQWVINGQKVWTSLAHLSQWCFVLAR
Rv3560c|M.tuberculosis_H37Rv SDLASVATTAELD----------GDQWVINGQKVWTSLAHLSQWCFVLAR
MMAR_5049|M.marinum_M SDLANVATTAELD----------GDQWVINGQKVWTSLAHLSQWCFVVAR
MUL_4125|M.ulcerans_Agy99 SDLANVATTAELD----------GDQWVINGQKVWTSLAHLSQWCFVVAR
MAB_0597|M.abscessus_ATCC_1997 SDLANVSTSAVLD----------GGDWVINGQKVWTSLALQSQWCFVVAR
TH_4120|M.thermoresistible__bu SDLASLKTRAVID----------GDELVITGQKIWTSKATYANRIYLLAR
MAV_2593|M.avium_104 SDLSSLSTRAVRD----------GDVYRVTGQKVWTTMADRADWMFALVR
MLBr_00737|M.leprae_Br4923 SDAASMRTRAKAD----------GDDWILNGFKCWITNGGKSTWYTVMAV
** :.: * * . :.* * * : . : :.
Mflv_1481|M.gilvum_PYR-GCK TEKGSKRHAGLSYLLVPLQQPGVEIRPIIQLTGD--SEFNEVFFDDARTA
Mvan_5285|M.vanbaalenii_PYR-1 TEKGSKRHAGLSYLLVPLQQPGVEIRPIIQLTGD--SEFNEVFFDDARTD
MSMEG_6012|M.smegmatis_MC2_155 TEKGSKRHAGLSYLLVPLDQPGVEIRPINQLTGD--SEFNEVFFDDARTE
Mb3590c|M.bovis_AF2122/97 TEKGSQRHAGLSYLLVPLDQPGVQIRPIVQITGT--AEFNEVFFDDARTD
Rv3560c|M.tuberculosis_H37Rv TEKGSQRHAGLSYLLVPLDQPGVQIRPIVQITGT--AEFNEVFFDDARTD
MMAR_5049|M.marinum_M TEKGSKRHAGLSYLLVPLDQPGVQIRPIVQITGT--AEFNEVFFDDARTD
MUL_4125|M.ulcerans_Agy99 TEKGSKRHAGLSYLLVPLDQPSVQIRPIVQITGT--AEFNEVFFDDARTD
MAB_0597|M.abscessus_ATCC_1997 TEPGSSRHKGLSYLLVPLDQPGVEIRPIIQLTGT--SEFNEVFFDNARTS
TH_4120|M.thermoresistible__bu TDPDAPKREGISYILADLNTPGIEVRPIRQITGA--SEFCEVFLDSVRVP
MAV_2593|M.avium_104 TGSGGKPSDGITYLLIPMKSPGITVRPLRDISGA--AHFAEVFLDDVAVP
MLBr_00737|M.leprae_Br4923 TDPD-KGANGISAFIVHKDDEGFSIGPKEKKLGIKGSPTTELYFDKCRIP
* . *:: :: . .. : * . * : *:::*.
Mflv_1481|M.gilvum_PYR-GCK ADLVVGEPGDGWRVAMGTLTFERGVSTLGQQIRYAREHSNLVELAE----
Mvan_5285|M.vanbaalenii_PYR-1 ADLVVGEPGDGWRVAMGTLTFERGVSTLGQQIRYAREHSNLVELAK----
MSMEG_6012|M.smegmatis_MC2_155 AGLVVGQPGDGWRVAMGTLTFERGVSTLGQQIRYAREHSNLVELAK----
Mb3590c|M.bovis_AF2122/97 ADLVVGAPGDGWRVAMATLTFERGVSTLGQQIVYARELSNLVELAR----
Rv3560c|M.tuberculosis_H37Rv ADLVVGAPGDGWRVAMATLTFERGVSTLGQQIVYARELSNLVELAR----
MMAR_5049|M.marinum_M ADLVVGAPGDGWRVAMGTLTFERGVSTLGQQIVYARELSDLAALAR----
MUL_4125|M.ulcerans_Agy99 ADLVVGAPGDGWRVAMGTLTFERGVSTLGQQIVYARELSDLAALAR----
MAB_0597|M.abscessus_ATCC_1997 ADLVVGEPGDGWRVAMGTLTFERGVSTLGNQITYARELSRLAELAK----
TH_4120|M.thermoresistible__bu LANVLGPLHGGWRVAKSTLGYER--VGQSRTHRIERRLGILVKMAQEPNA
MAV_2593|M.avium_104 VENVVGDEGAGWSIMRTSLGHERATAFLADEFKYRRTADRVIDLVV----
MLBr_00737|M.leprae_Br4923 GDRIIGEPGTGFKTALATLDHTRPTIGAQAVGIAQGALDAAIVYTKDRKQ
::* *: :* . * . .
Mflv_1481|M.gilvum_PYR-GCK -KTGAADDPLIRERLVRSWAGLKAMRSY--ALATMD---VEKPG-QDNIS
Mvan_5285|M.vanbaalenii_PYR-1 -RTGAADDPLIRERLTRSWAGLKAMRSY--ALATMD---VEQPG-MDNVS
MSMEG_6012|M.smegmatis_MC2_155 -RTGAADDPLIRERLVQSWTGLQAMRSY--ALATMD---VEQPG-QDNVS
Mb3590c|M.bovis_AF2122/97 -RTAAADDPLIRERLTRAWTGLRAMRSY--ALATMEGPAVEQPG-QDNVS
Rv3560c|M.tuberculosis_H37Rv -RTAAADDPLIRERLTRAWTGLRAMRSY--ALATMEGPAVEQPG-QDNVS
MMAR_5049|M.marinum_M -RTGAADDPFIRERLTRAWTGLRAMRSY--ALATMD---VEQPG-QDNVS
MUL_4125|M.ulcerans_Agy99 -RTGAADDPFIREHLTRAWTGLRAMRSY--ALATMD---VEQPG-QDNVS
MAB_0597|M.abscessus_ATCC_1997 -SNGAADDPAIRERLAQAYVGMRTMRAY--ALATMD---EERPG-QDNVS
TH_4120|M.thermoresistible__bu LTDSGFDDSYVADRIVRYAAQVEALRQIA-AQATAAGVRGVAPGPEASVA
MAV_2593|M.avium_104 -AQRLDDDPLVRQDVARLESGVRTIAANS-ARALAAVLRGEDPGGMASVN
MLBr_00737|M.leprae_Br4923 FGESISTFQSIQFMLADMAMKVEAARLIVYAAAARAERGEPDLGFISAAS
: :. :.: * * *
Mflv_1481|M.gilvum_PYR-GCK KLLWANWHRELGEIAMDIEGAAGLTLP----DGEFSEWQRLYLFSRSDTI
Mvan_5285|M.vanbaalenii_PYR-1 KLLWANWHRELGEIAMDVQGMAGLTLP----GGEFDEWQRLYLFSRSDTI
MSMEG_6012|M.smegmatis_MC2_155 KLLWANWHRELGEIAMDVQGMAGLTLE----NGEFDEWQRLYLFSRADTI
Mb3590c|M.bovis_AF2122/97 KLLWANWHRNLGELAMDVIGKPGMTMP----DGEFDEWQRLYLFTRADTI
Rv3560c|M.tuberculosis_H37Rv KLLWANWHRNLGELAMDVIGKPGMTMP----DGEFDEWQRLYLFTRADTI
MMAR_5049|M.marinum_M KLLWANWHRSLGELAMDVIGKPGLTLA----DGEFDEWQRLFLFTRADTI
MUL_4125|M.ulcerans_Agy99 KLLWANWHRSLGELAMDVIGKPGLTLA----DGEFDEWQRLFLFTRADTI
MAB_0597|M.abscessus_ATCC_1997 KLLWANWHRDLGELAMEIIGTSTLLTN----AGEMDEWQRLFLFSRADTI
TH_4120|M.thermoresistible__bu KLLTSEVDQSMANFGVELAGAPGVLERGSPGAAKRGNVAASYLLMRAATF
MAV_2593|M.avium_104 RLVKSEFEQHMHALALRAAGPYAALGSRAPDAVDKGRWTFGYLMSRATTI
MLBr_00737|M.leprae_Br4923 KCFASDIAMEVTTDAVQLFGGAGYTSD--------FPVERFMRDAKITQI
: . :: : .: * : :
Mflv_1481|M.gilvum_PYR-GCK YGGSNEIQRNIIAERVLGLPREARH-------------------------
Mvan_5285|M.vanbaalenii_PYR-1 YGGSNEIQRNIIAERVLGLPREAKG-------------------------
MSMEG_6012|M.smegmatis_MC2_155 YGGSNEIQRNIIAERVLGLPREVKG-------------------------
Mb3590c|M.bovis_AF2122/97 YGGSNEIQRNIIAERVLGLPREAKG-------------------------
Rv3560c|M.tuberculosis_H37Rv YGGSNEIQRNIIAERVLGLPREAKG-------------------------
MMAR_5049|M.marinum_M YGGSNEIQRNIIAERVLGLPREAKG-------------------------
MUL_4125|M.ulcerans_Agy99 YGGSNEIQRNIIAERVLGLPREAKG-------------------------
MAB_0597|M.abscessus_ATCC_1997 YGGSNEIQRNIISERVLGLPREAR--------------------------
TH_4120|M.thermoresistible__bu GAGTSEIQRNVIAERLLGLPRDN---------------------------
MAV_2593|M.avium_104 GAGTAEIQRNTIAESVLGLPSHRGEGTREAAVTPGAPLAVPEEDERELRE
MLBr_00737|M.leprae_Br4923 YEGTNQIQRVVMSRALLR--------------------------------
*: :*** ::. :*
Mflv_1481|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5285|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_6012|M.smegmatis_MC2_155 --------------------------------------------------
Mb3590c|M.bovis_AF2122/97 --------------------------------------------------
Rv3560c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_5049|M.marinum_M --------------------------------------------------
MUL_4125|M.ulcerans_Agy99 --------------------------------------------------
MAB_0597|M.abscessus_ATCC_1997 --------------------------------------------------
TH_4120|M.thermoresistible__bu --------------------------------------------------
MAV_2593|M.avium_104 VLAATLQAKVDEAALLDRKRPVDAAEPEVWSALVEFGLPGLAVDETLGGA
MLBr_00737|M.leprae_Br4923 --------------------------------------------------
Mflv_1481|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5285|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_6012|M.smegmatis_MC2_155 --------------------------------------------------
Mb3590c|M.bovis_AF2122/97 --------------------------------------------------
Rv3560c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_5049|M.marinum_M --------------------------------------------------
MUL_4125|M.ulcerans_Agy99 --------------------------------------------------
MAB_0597|M.abscessus_ATCC_1997 --------------------------------------------------
TH_4120|M.thermoresistible__bu --------------------------------------------------
MAV_2593|M.avium_104 GARPRLLYTAVEEAAKALAPAPLVPTVIGLDVALHCGAKALVQRITGGAT
MLBr_00737|M.leprae_Br4923 --------------------------------------------------
Mflv_1481|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5285|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_6012|M.smegmatis_MC2_155 --------------------------------------------------
Mb3590c|M.bovis_AF2122/97 --------------------------------------------------
Rv3560c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_5049|M.marinum_M --------------------------------------------------
MUL_4125|M.ulcerans_Agy99 --------------------------------------------------
MAB_0597|M.abscessus_ATCC_1997 --------------------------------------------------
TH_4120|M.thermoresistible__bu --------------------------------------------------
MAV_2593|M.avium_104 AAFAVPVNDSGWVTSGPELPEWDGTGLSGVVPIVAGAPHAEVLVALARVA
MLBr_00737|M.leprae_Br4923 --------------------------------------------------
Mflv_1481|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5285|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_6012|M.smegmatis_MC2_155 --------------------------------------------------
Mb3590c|M.bovis_AF2122/97 --------------------------------------------------
Rv3560c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_5049|M.marinum_M --------------------------------------------------
MUL_4125|M.ulcerans_Agy99 --------------------------------------------------
MAB_0597|M.abscessus_ATCC_1997 --------------------------------------------------
TH_4120|M.thermoresistible__bu --------------------------------------------------
MAV_2593|M.avium_104 AGVGEVLVAVDASADGVEVTAQQPLDLTGTVGAVTFSGAAGEVLADGTDV
MLBr_00737|M.leprae_Br4923 --------------------------------------------------
Mflv_1481|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5285|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_6012|M.smegmatis_MC2_155 --------------------------------------------------
Mb3590c|M.bovis_AF2122/97 --------------------------------------------------
Rv3560c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_5049|M.marinum_M --------------------------------------------------
MUL_4125|M.ulcerans_Agy99 --------------------------------------------------
MAB_0597|M.abscessus_ATCC_1997 --------------------------------------------------
TH_4120|M.thermoresistible__bu --------------------------------------------------
MAV_2593|M.avium_104 PRMLAAARRRAALAVAADSVGVASRALAMAVQWARERHQFGRPIGSFQAV
MLBr_00737|M.leprae_Br4923 --------------------------------------------------
Mflv_1481|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5285|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_6012|M.smegmatis_MC2_155 --------------------------------------------------
Mb3590c|M.bovis_AF2122/97 --------------------------------------------------
Rv3560c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_5049|M.marinum_M --------------------------------------------------
MUL_4125|M.ulcerans_Agy99 --------------------------------------------------
MAB_0597|M.abscessus_ATCC_1997 --------------------------------------------------
TH_4120|M.thermoresistible__bu --------------------------------------------------
MAV_2593|M.avium_104 SHRCADMLVALEGARSQILEAAESDLEESGYLADLAAAAALDAGAAATEG
MLBr_00737|M.leprae_Br4923 --------------------------------------------------
Mflv_1481|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5285|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_6012|M.smegmatis_MC2_155 --------------------------------------------------
Mb3590c|M.bovis_AF2122/97 --------------------------------------------------
Rv3560c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_5049|M.marinum_M --------------------------------------------------
MUL_4125|M.ulcerans_Agy99 --------------------------------------------------
MAB_0597|M.abscessus_ATCC_1997 --------------------------------------------------
TH_4120|M.thermoresistible__bu --------------------------------------------------
MAV_2593|M.avium_104 ALQIHGGIGFTWEHPIHLLLRRARANSVLVGRADALRDRAAGELLAPYRQ
MLBr_00737|M.leprae_Br4923 --------------------------------------------------
Mflv_1481|M.gilvum_PYR-GCK ----
Mvan_5285|M.vanbaalenii_PYR-1 ----
MSMEG_6012|M.smegmatis_MC2_155 ----
Mb3590c|M.bovis_AF2122/97 ----
Rv3560c|M.tuberculosis_H37Rv ----
MMAR_5049|M.marinum_M ----
MUL_4125|M.ulcerans_Agy99 ----
MAB_0597|M.abscessus_ATCC_1997 ----
TH_4120|M.thermoresistible__bu ----
MAV_2593|M.avium_104 QRSE
MLBr_00737|M.leprae_Br4923 ----