For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SSILPGAAAIVGIGQTEFSKESGRSELQLACEAVSAALDDAGLTPADVDGMVTFTMDSSDEIDIARNVGI GDLSFFSRVHHGGGAAAGTVVHAAMAVATGVADVVVCWRAFNERSGFRFGGSGRNMAETPLFMAHYAPFG LLTPAAWVALHAQRYMSTYGVTNEDFGRIAVVDRKHAATNPDAWFYQRPITLEDHQNSRWIVEPVLRLLD CCQESDGGVALVVTSVERARDLRRPPAVIAAAAQGAAADGEMMTSYYRDDITGLPEMGVVARQLWRDSGC KPQDIQTAFIYDHFTPFVFVQLEELGFCGRGEAKDFATVERLSLGGEFPINTNGGLLGEAYIHGMNGITE AVRQVRGTSHNQVADVEHVLVTSGTGVPTSGLILAPAG*
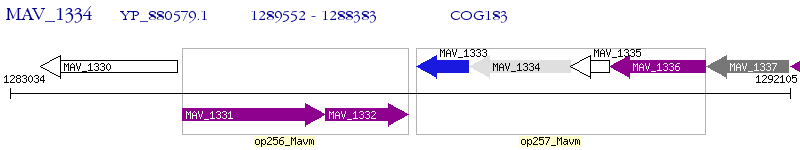
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_1334 | - | - | 100% (389) | lipid-transfer protein |
| M. avium 104 | MAV_0621 | - | e-145 | 67.44% (390) | lipid-transfer protein |
| M. avium 104 | MAV_0477 | - | 2e-71 | 41.98% (393) | lipid-transfer protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3570c | ltp2 | 1e-144 | 66.32% (386) | lipid-transfer protein |
| M. gilvum PYR-GCK | Mflv_1513 | - | 1e-142 | 67.36% (386) | lipid-transfer protein |
| M. tuberculosis H37Rv | Rv3540c | ltp2 | 1e-144 | 66.32% (386) | lipid-transfer protein |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_0618 | - | 1e-140 | 65.71% (385) | lipid-transfer protein |
| M. marinum M | MMAR_4250 | ltp2 | 0.0 | 90.39% (385) | keto acyl-CoA thiolase, Ltp2 |
| M. smegmatis MC2 155 | MSMEG_5990 | - | 1e-141 | 66.23% (385) | lipid-transfer protein |
| M. thermoresistible (build 8) | TH_0556 | ltp2 | 0.0 | 85.12% (383) | PROBABLE LIPID TRANSFER PROTEIN OR KETO ACYL-COA THIOLASE |
| M. ulcerans Agy99 | MUL_0935 | ltp2 | 0.0 | 89.87% (385) | lipid-transfer protein |
| M. vanbaalenii PYR-1 | Mvan_5253 | - | 1e-145 | 68.13% (386) | lipid-transfer protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1513|M.gilvum_PYR-GCK ---MLSGKAAIAGIGATDFSKNSGRSELRLAAEAVLDALDDAGLTTADVD
Mvan_5253|M.vanbaalenii_PYR-1 ---MLSGKAAIAGIGATDFSKNSGRSELRLAAEAVLDALDDAGLTPSDVD
MAB_0618|M.abscessus_ATCC_1997 -MTGLSGRSAIAGIGATDFSKKSGRSELRLAAEAVLDALDDAGLSPSDVD
MSMEG_5990|M.smegmatis_MC2_155 -MTGLSGKAAIAGIGATDFSKNSGRSELRLAAECVLDALDDAGLEPSDVD
Mb3570c|M.bovis_AF2122/97 ---MLSGQAAIVGIGATDFSKNSGRSELRLAAEAVLDALADAGLSPTDVD
Rv3540c|M.tuberculosis_H37Rv ---MLSGQAAIVGIGATDFSKNSGRSELRLAAEAVLDALADAGLSPTDVD
MMAR_4250|M.marinum_M MTAPLAGAAAIVGIGQTEFSKESGRSELQLACEAVSAALDDAGVTPGEVD
MUL_0935|M.ulcerans_Agy99 MTAPLAGAAAIVGIGQTEFSKESGRSELQLACEAVSAALDDAGVTPGEVD
MAV_1334|M.avium_104 MSSILPGAAAIVGIGQTEFSKESGRSELQLACEAVSAALDDAGLTPADVD
TH_0556|M.thermoresistible__bu MTG-ISGRAAIVGIGQTEFSKESGRSEMQLACEAVKAALDDAGLTPADVD
:.* :**.*** *:***:*****::**.*.* ** ***: . :**
Mflv_1513|M.gilvum_PYR-GCK GLVTFSMDSNLETAVARSTGIGELKFFSQIGYGGGAAAATVQQAALAVAT
Mvan_5253|M.vanbaalenii_PYR-1 GLVTFTMDSNLETAVARSTGIGDLTFFSQIGYGGGAAAATVQQAALAVAT
MAB_0618|M.abscessus_ATCC_1997 GLVTFTMDSNLETAVARATGIGELKFFSQIGYGGGAAAATVQQAALAVAA
MSMEG_5990|M.smegmatis_MC2_155 GLVTFTMDSNQETAVARSTGIGELKFFSQIGYGGGAAAATVQQAALAVAT
Mb3570c|M.bovis_AF2122/97 GLTTFTMDTNTEIAVARAAGIGELTFFSKIHYGGGAACATVQHAAMAVAT
Rv3540c|M.tuberculosis_H37Rv GLTTFTMDTNTEIAVARAAGIGELTFFSKIHYGGGAACATVQHAAMAVAT
MMAR_4250|M.marinum_M GMVTFTMDASDEIDVARNVGIGDLNFFSRVPHGGGAAAGTVAHAAMAVAT
MUL_0935|M.ulcerans_Agy99 GMVTFTMDASDEIDVARNVGIGDLNFFSRVPHGGGAAAGTVAHAAMAVAT
MAV_1334|M.avium_104 GMVTFTMDSSDEIDIARNVGIGDLSFFSRVHHGGGAAAGTVVHAAMAVAT
TH_0556|M.thermoresistible__bu GMVTFTVDENEEIEVARNVGIGNLTFFTRVPHGGGAAAGTVLQAAMAVAT
*:.**::* . * :** .***:*.**::: :*****..** :**:***:
Mflv_1513|M.gilvum_PYR-GCK GVAEVVVAYRAFNERSEHRFGQVMTGLTVNADSRGVEYSWSYPHGLSTPA
Mvan_5253|M.vanbaalenii_PYR-1 GVAEVVVAYRAFNERSEHRFGQVMTGLTVNADSRGVEYSWSYPHGLSTPA
MAB_0618|M.abscessus_ATCC_1997 GVAEVVVAYRAFNERSEFRFGQVMTGLSSTADSRGVEYSWSYPHGLSTPA
MSMEG_5990|M.smegmatis_MC2_155 GVAEVVVAYRAFNERSEFRFGQVMTGLTTNADSRGVEYSWSYPHGLSTPA
Mb3570c|M.bovis_AF2122/97 GVADVVVAYRAFNERSGMRFGQVQTRLTENADSTGVDNSFSYPHGLSTPA
Rv3540c|M.tuberculosis_H37Rv GVADVVVAYRAFNERSGMRFGQVQTRLTENADSTGVDNSFSYPHGLSTPA
MMAR_4250|M.marinum_M GVAEVVVCYRAFNERSGMRFGGSGRTSTETP----LFMAHYAPFGLLTPA
MUL_0935|M.ulcerans_Agy99 GVAEVVVCYRAFNERSGMRFGGSGRTSTETP----LFMAHYAPFGLLTPA
MAV_1334|M.avium_104 GVADVVVCWRAFNERSGFRFGGSGRNMAETP----LFMAHYAPFGLLTPA
TH_0556|M.thermoresistible__bu GVADVVVCYRAFNERSGFRFGGAGRKPTAPP----LWLAPYAPFGLMTPA
***:***.:******* *** : . : : *.** ***
Mflv_1513|M.gilvum_PYR-GCK ASVAMIAQRYMHEFGATSADFGVISVADRKHAANNPKAHFYGKPITIEDH
Mvan_5253|M.vanbaalenii_PYR-1 ASVAMIARRYMHEYGATSADFGAVSVADRKHAATNPKAHFYGKPITIEDH
MAB_0618|M.abscessus_ATCC_1997 ASVAMIARRYMHEYGATSADFGAVSVADRKHAATNPKAFFYGKPITIEDH
MSMEG_5990|M.smegmatis_MC2_155 ASVAMIARRYMHEYGATSADFGVVSVADRKHAANNPKAHFYGKPITIEDH
Mb3570c|M.bovis_AF2122/97 AQVAMIARRYMHLSGATSRDFGAVSVADRKHAANNPKAYFYGKPITIEDH
Rv3540c|M.tuberculosis_H37Rv AQVAMIARRYMHLSGATSRDFGAVSVADRKHAANNPKAYFYGKPITIEDH
MMAR_4250|M.marinum_M AWVALHAQRYMSTYGVTNEDFGRIAVVDRAHAARNPDAWFYQRPITLADH
MUL_0935|M.ulcerans_Agy99 AWVALHAQRYMSTYGVTNEDFGRIAVVDRAHAARNPDAWFYQRPITLADH
MAV_1334|M.avium_104 AWVALHAQRYMSTYGVTNEDFGRIAVVDRKHAATNPDAWFYQRPITLEDH
TH_0556|M.thermoresistible__bu GWVALHAQRYMTTYGVTNEDFGKISVIDRKHAANNPDAWFYQRPITLEDH
. **: *:*** *.*. *** ::* ** *** **.* ** :***: **
Mflv_1513|M.gilvum_PYR-GCK QNSRWIAEP-LRLLDCCQETDGGVAIVVTTPERARDLKHRPAIIEAAAQG
Mvan_5253|M.vanbaalenii_PYR-1 QNSRWIAEP-LRLLDCCQETDGGVAIVVTTPERAKDLRHRPAVIEAAAQG
MAB_0618|M.abscessus_ATCC_1997 QNSRMIADP-VRLLDCCQESDGGVAIVVTTPERAKDLKNRPVIIEAAAQG
MSMEG_5990|M.smegmatis_MC2_155 QNSRWIAEP-LRLLDCCQETDGGVAIVVTTPERAKDLKHRPVTIEAAAQG
Mb3570c|M.bovis_AF2122/97 QNSRWIAEP-LRLLDCCQETDGAVAIVVTSAARARDLKQRPVVIEAAAQG
Rv3540c|M.tuberculosis_H37Rv QNSRWIAEP-LRLLDCCQETDGAVAIVVTSAARARDLKQRPVVIEAAAQG
MMAR_4250|M.marinum_M QNSRWIIEPVLRLLDCCQESDGGVALVVTSTERARDLRQPAAVITAAAQG
MUL_0935|M.ulcerans_Agy99 QNSRWIIEPVLRLLDCCQESDGGVALVVTSTERARDLRQPAAVIPAAAQR
MAV_1334|M.avium_104 QNSRWIVEPVLRLLDCCQESDGGVALVVTSVERARDLRRPPAVIAAAAQG
TH_0556|M.thermoresistible__bu QNSRWIIEPVLRLLDCCQESDGGVALVVTSVERARDLRHRPAVITAAAQG
**** * :* :********:**.**:***: **:**:. .. * ****
Mflv_1513|M.gilvum_PYR-GCK AGADQFTMYSYYREELG-LPEMGLVGRQLWAQSGLTPQDIQTAVLYDHFT
Mvan_5253|M.vanbaalenii_PYR-1 AGADQFTMYSYYRDELG-LPEMGLVGRQLWEQSGLSPEDIQTAILYDHFT
MAB_0618|M.abscessus_ATCC_1997 SGEDQFTMYSYYRDELG-LPEMGLVGRQLWDQSGLTPNDIQTAILYDHFT
MSMEG_5990|M.smegmatis_MC2_155 SGADQFTMYSYYRDELG-LPEMGLVGRQLWQQSGLTPDDIQTAILYDHFT
Mb3570c|M.bovis_AF2122/97 CSPDQYTMVSYYRPELDGLPEMGLVGRQLWAQSGLTPADVQTAVLYDHFT
Rv3540c|M.tuberculosis_H37Rv CSPDQYTMVSYYRPELDGLPEMGLVGRQLWAQSGLTPADVQTAVLYDHFT
MMAR_4250|M.marinum_M AAAEGEMMTSYYREDITGLPEMGVVAKRLWRDSGLKPADIQTAFIYDHFT
MUL_0935|M.ulcerans_Agy99 AAAEGEMMTSYYREDITGLPVMGVVAKRLWRDSGLKPADIQTAFIYDHFT
MAV_1334|M.avium_104 AAADGEMMTSYYRDDITGLPEMGVVARQLWRDSGCKPQDIQTAFIYDHFT
TH_0556|M.thermoresistible__bu AAYDGEVMTSYYRDDITGLPEMGVVAEKLWRDSGLKPSDISTAFLYDHFT
.. : * **** :: ** **:*..:** :** .* *:.**.:*****
Mflv_1513|M.gilvum_PYR-GCK PYTLIQLEELGFCGKGEAKDFIAGGAIEIGGRLPINTHGGQLGEAYIHGM
Mvan_5253|M.vanbaalenii_PYR-1 PYTLIQLEELGFCGKGEAKDFIAGGAIEIGGRLPINTHGGQLGEAYIHGM
MAB_0618|M.abscessus_ATCC_1997 PYTLLQLEELGFCGKGEAKDFIANGAIELGGKLPINTHGGQLGEAYIHGM
MSMEG_5990|M.smegmatis_MC2_155 PYTLLQLEELGFCGKGEAKDFIAGGAIEIGGKLPINTHGGQLGEAYIHGM
Mb3570c|M.bovis_AF2122/97 PFTLIQLEELGFCGKGEAKDFIADGAIEVGGRLPINTHGGQLGEAYIHGM
Rv3540c|M.tuberculosis_H37Rv PFTLIQLEELGFCGKGEAKDFIADGAIEVGGRLPINTHGGQLGEAYIHGM
MMAR_4250|M.marinum_M PFVFTQLEELGFCGRGEAKEFATVENLSLGGLLPINTNGGLLGEAYIHGM
MUL_0935|M.ulcerans_Agy99 PFVFTQLEELGFCGRGEAKEFATVENLSLGGLLPINTNGGLLGEAYIHGM
MAV_1334|M.avium_104 PFVFVQLEELGFCGRGEAKDFATVERLSLGGEFPINTNGGLLGEAYIHGM
TH_0556|M.thermoresistible__bu PFVFTQLEELGFCGRGEAKDFVSVDELSLGGSLPINTNGGLLGEAYIHGM
*:.: *********:****:* : :.:** :****:** *********
Mflv_1513|M.gilvum_PYR-GCK NGIAEGVRQLRGTSVNQVPDVEHVLVTAGTGVPTSGLILG---
Mvan_5253|M.vanbaalenii_PYR-1 NGIAEGVRQLRGTSVNQVPDVEHVLVTAGTGVPTSGLILG---
MAB_0618|M.abscessus_ATCC_1997 NGIAEGVRQLRGTSVNQVPNVEHVLVTAGTGVPTSGLILG---
MSMEG_5990|M.smegmatis_MC2_155 NGIAEGVRQLRGTSVNQVEGVEHVLVTAGTGVPTSGLILG---
Mb3570c|M.bovis_AF2122/97 NGIAEGVRQLRGTSVNPVAGVEHVLVTAGTGVPTSGLILG---
Rv3540c|M.tuberculosis_H37Rv NGIAEGVRQLRGTSVNPVAGVEHVLVTAGTGVPTSGLILG---
MMAR_4250|M.marinum_M NGITEAVRQVRGTSYNQVDHVEHVLVTSGTGVPTSGLILAPAG
MUL_0935|M.ulcerans_Agy99 NGITEAVRQVRGTSYNQVDHVEHVLVTSGTGVPTSGLILAPAG
MAV_1334|M.avium_104 NGITEAVRQVRGTSHNQVADVEHVLVTSGTGVPTSGLILAPAG
TH_0556|M.thermoresistible__bu NGITEGVRQVRGTSYNQVENVEHVLVTSGTGVPTSGLILAPG-
***:*.***:**** * * *******:***********.