For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTTEKRTAGWAASVVAGGALAMSVVGLAGGPVAAAAPAPAPTGHWCPGDPWNPSWGNVLDWDWHQCHDWQ HPGGPTGPAGWGPWGPPPAWAPPPPPQPAWAPGAQMMWNPTGAGNWGIWNNGVWTPL
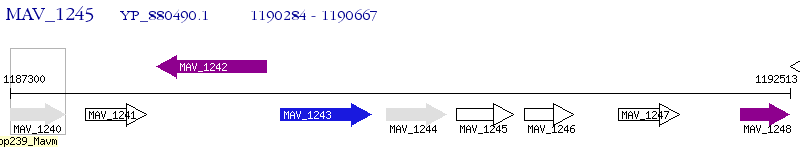
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_1245 | - | - | 100% (127) | secreted protein antigen |
| M. avium 104 | MAV_0920 | - | 9e-06 | 36.67% (60) | hypothetical protein MAV_0920 |
| M. avium 104 | MAV_3538 | - | 4e-05 | 33.68% (95) | hypothetical protein MAV_3538 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3341c | - | 4e-13 | 42.22% (90) | secreted protein antigen |
| M. gilvum PYR-GCK | Mflv_3438 | - | 9e-07 | 31.25% (112) | hypothetical protein Mflv_3438 |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_0269c | - | 2e-11 | 40.21% (97) | hypothetical protein MAB_0269c |
| M. marinum M | MMAR_3811 | - | 1e-12 | 42.45% (106) | hypothetical protein MMAR_3811 |
| M. smegmatis MC2 155 | MSMEG_3545 | - | 2e-10 | 27.23% (202) | hypothetical protein MSMEG_3545 |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_3736 | - | 6e-13 | 42.45% (106) | hypothetical protein MUL_3736 |
| M. vanbaalenii PYR-1 | Mvan_2955 | - | 4e-11 | 36.63% (101) | hypothetical protein Mvan_2955 |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_3811|M.marinum_M -------------MKTYPRLAATGLAMVLGLGLAGVGVASEAAAQPG-AP
MUL_3736|M.ulcerans_Agy99 -----------MPMKTYPRLAATGLAMVLGLGLAGVGVASEAAAQPG-AP
MAB_0269c|M.abscessus_ATCC_199 -------------MKILYGIAASAAIAASGLGLAATANAAPAPALPS-AP
Mb3341c|M.bovis_AF2122/97 -------------MYRFACRTLMLAACILATGVAGLGVGAQSAAQTAPVP
MSMEG_3545|M.smegmatis_MC2_155 -MMIVKTTTTRFVAAALTVGALGFGATAFGTPVASAGPAAPVPLKPGDGG
MAV_1245|M.avium_104 ---------MTTEKRTAGWAASVVAGGALAMSVVGLAGGPVAAAAPAPAP
Mflv_3438|M.gilvum_PYR-GCK -------------MALLKTWMKTALPAAVMAGGLAAAVLGSGAAPAFAQP
Mvan_2955|M.vanbaalenii_PYR-1 MSAVRNDRGSSPGIGVLTMKLTHVLASAAVFGGLAAGAMG-VASTAAAQP
. . . .
MMAR_3811|M.marinum_M T-QWCPGDFWDP-------GWG--QNWDMGHCH----------DNWRGPG
MUL_3736|M.ulcerans_Agy99 T-QWCPGDFWDP-------AWG--QNWDMGHCH----------DNWRGPG
MAB_0269c|M.abscessus_ATCC_199 TYHWCPGEFWNP-------IWG--FNMNWGECHADGILDRDRPDDWRGDD
Mb3341c|M.bovis_AF2122/97 DYYWCPGQPFDP-------AWG--PNWDPYTCH----------DDFHRDS
MSMEG_3545|M.smegmatis_MC2_155 CFPFCDGGPGKPGGPGGPGHGG--PGGPGGPGHGG----PGGPGGHEGPG
MAV_1245|M.avium_104 TGHWCPGDPWNP-------SWGNVLDWDWHQCH-----------DWQHPG
Mflv_3438|M.gilvum_PYR-GCK --APPPAP-VGE----------QPPAWAPPKPAE----------IWN-GH
Mvan_2955|M.vanbaalenii_PYR-1 GWTPPPTPPAGPN-------TGQPPAWAPPKPVDP---------LWANGQ
MMAR_3811|M.marinum_M PNPQPGHAGPG----GPGEPAPG-QQGPG-GPGGPGGPGEGPGGPGGPGE
MUL_3736|M.ulcerans_Agy99 PNPQPGHAGPG----GPGEPAPG-QQGPG-GPGGP---------------
MAB_0269c|M.abscessus_ATCC_199 GRWRNNGPGPG----DNRGPDRGPDRGPDRGPGND---------------
Mb3341c|M.bovis_AF2122/97 DGPDHSRDYPGPILEGPVLDDPGAAPPPPAAGGGA---------------
MSMEG_3545|M.smegmatis_MC2_155 GPGRHDGPGPG----GPGGPGRHDGPGGPGGPGWHDPGHGPDKGPWWANN
MAV_1245|M.avium_104 GPTGPAGWGPWG---PPPAWAPPPPPQPAWAPGAQMMWNPTGAGNWGIWN
Mflv_3438|M.gilvum_PYR-GCK PVVWNSAWGGRWGAWINGGFISLTSNPVTGGG------------------
Mvan_2955|M.vanbaalenii_PYR-1 PQVWDQGWQ-HWGVWLNGVFVPTY--------------------------
MMAR_3811|M.marinum_M GPGGPGGPH-----------------------------------------
MUL_3736|M.ulcerans_Agy99 --------H-----------------------------------------
MAB_0269c|M.abscessus_ATCC_199 -------WHR----------------------------------------
Mb3341c|M.bovis_AF2122/97 --------------------------------------------------
MSMEG_3545|M.smegmatis_MC2_155 RHDWWDDRQGPPPWGWGPPPPVHWNGGPLPHTINYWGYNLNPVWHDGFRQ
MAV_1245|M.avium_104 NGVWTPL-------------------------------------------
Mflv_3438|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_2955|M.vanbaalenii_PYR-1 --------------------------------------------------
MMAR_3811|M.marinum_M ------------------
MUL_3736|M.ulcerans_Agy99 ------------------
MAB_0269c|M.abscessus_ATCC_199 ------------------
Mb3341c|M.bovis_AF2122/97 ------------------
MSMEG_3545|M.smegmatis_MC2_155 WGVWLFGIWIPIFGVGFN
MAV_1245|M.avium_104 ------------------
Mflv_3438|M.gilvum_PYR-GCK ------------------
Mvan_2955|M.vanbaalenii_PYR-1 ------------------