For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
PPAPPDWYQPPPPYAPSFAGQDVPPPPPRPWWSPNEPMWSVRFHQWGTYVAGTWVPF*
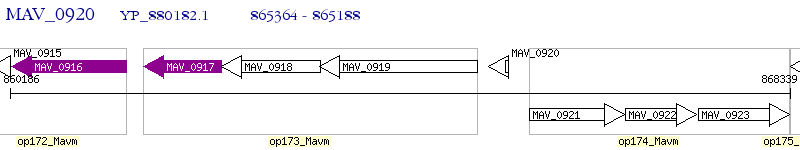
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_0920 | - | - | 100% (58) | hypothetical protein MAV_0920 |
| M. avium 104 | MAV_1245 | - | 6e-06 | 36.67% (60) | secreted protein antigen |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | - | - | - | - | - |
| M. smegmatis MC2 155 | MSMEG_5916 | - | 6e-06 | 50.00% (34) | hypothetical protein MSMEG_5916 |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_2955 | - | 4e-06 | 36.21% (58) | hypothetical protein Mvan_2955 |
CLUSTAL 2.0.9 multiple sequence alignment
MAV_0920|M.avium_104 -------------------------------------------MPPAPPD
Mvan_2955|M.vanbaalenii_PYR-1 MSAVRNDRGSSPGIGVLTMKLTHVLASAAVFGGLAAGAMGVASTAAAQPG
MSMEG_5916|M.smegmatis_MC2_155 ---------------MGGALFAGATGAAALAGAQPAGDSAPMSVLGPVPA
. *
MAV_0920|M.avium_104 WYQPPPPYAPSFAGQD---VPPPPPRPWWSPNEP--MWSVR----FHQWG
Mvan_2955|M.vanbaalenii_PYR-1 WTPPPTPPAGPNTGQPPAWAPPKPVDPLWANGQPQ-VWDQG----WQHWG
MSMEG_5916|M.smegmatis_MC2_155 FPPPPPPPP-PFWGPP---PPPPPPPPFWRPPPPPPIWLPPPPPPPPFWG
: **.* . . * ** * * * * :* **
MAV_0920|M.avium_104 ----TYVAGTWVPF-
Mvan_2955|M.vanbaalenii_PYR-1 ----VWLNGVFVPTY
MSMEG_5916|M.smegmatis_MC2_155 PPPPPRPPGHWR---
* :