For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
PRFDPLPERRPAIVAGASSGIGEATAIELAAHGFPVALGARRVEKLNDIVGKINADGGEAVGFHLDVTDP NSVKSFVAQAVDALGDIEVLVAGAGDTYFGKLAEIAGDEFESQLQIHLVGAFRLASAVLPGMLERQRGDL IFVGSDVALRQRPHMGAYGAAKAALVAMVNNFQMELEGTGVRASVVHPGPTKTAMGWSLPAEKIGPALED WAKWGQARHDYFLRAADLARAITFVAETPRGGFIANMELQPEAPLADNKDRQKLALGEEGMPGQ*
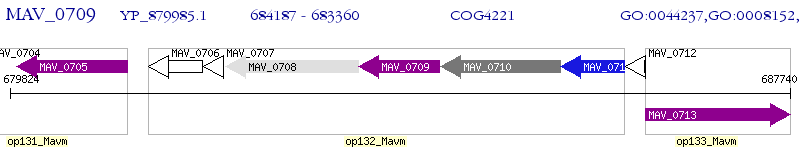
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_0709 | - | - | 100% (275) | short chain dehydrogenase |
| M. avium 104 | MAV_3227 | - | 2e-23 | 32.98% (191) | oxidoreductase, short-chain dehydrogenase/reductase family |
| M. avium 104 | MAV_5221 | - | 4e-23 | 34.92% (189) | NAD dependent epimerase/dehydratase family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0788c | - | 1e-112 | 72.73% (275) | short chain dehydrogenase |
| M. gilvum PYR-GCK | Mflv_1597 | - | 1e-134 | 84.98% (273) | short chain dehydrogenase |
| M. tuberculosis H37Rv | Rv0765c | - | 1e-112 | 72.73% (275) | short chain dehydrogenase |
| M. leprae Br4923 | MLBr_00429 | - | 4e-24 | 35.03% (197) | putative oxidoreductase |
| M. abscessus ATCC 19977 | MAB_1215c | - | 1e-110 | 69.34% (274) | short chain dehydrogenase |
| M. marinum M | MMAR_4931 | - | 1e-125 | 81.48% (270) | short-chain alcohol dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_5862 | - | 1e-127 | 81.09% (275) | short chain dehydrogenase |
| M. thermoresistible (build 8) | TH_1257 | - | 1e-122 | 80.74% (270) | short chain dehydrogenase |
| M. ulcerans Agy99 | MUL_0474 | - | 1e-122 | 80.00% (270) | short chain dehydrogenase |
| M. vanbaalenii PYR-1 | Mvan_5160 | - | 1e-129 | 83.46% (272) | short chain dehydrogenase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb0788c|M.bovis_AF2122/97 MPRFEPHPARRTTVVAGASSGIGAATATELAGRGFPVALGARRMDKLAEL
Rv0765c|M.tuberculosis_H37Rv MPRFEPHPARRTTVVAGASSGIGAATATELAGRGFPVALGARRMDKLAEL
MAB_1215c|M.abscessus_ATCC_199 MPRFEPHPARRPSIIAGASSGIGAATAIELAGRGFPVALGARRVDKLTEL
Mflv_1597|M.gilvum_PYR-GCK MPRFDPLPDRRPALVAGASSGIGEATAIRLAANGFPVALGARRVEKLEEI
Mvan_5160|M.vanbaalenii_PYR-1 MPRFDPLPDRRPALVAGASSGIGEATAIALAAHGFPVALGARRVEKCQEI
MAV_0709|M.avium_104 MPRFDPLPERRPAIVAGASSGIGEATAIELAAHGFPVALGARRVEKLNDI
MMAR_4931|M.marinum_M MPRFQPHPDRRPAIVAGASSGIGAATAVELAAHGFPVALGARRVQKCEEI
MUL_0474|M.ulcerans_Agy99 MPRFQPHPDRRPAIVAGASPGIGAATAVDLAAHGFPDALGARRVQKCEKI
TH_1257|M.thermoresistible__bu MPRFDPHPERRPAIVAGASSGIGAATAKELAARGFPVALGARRVEKCEEL
MSMEG_5862|M.smegmatis_MC2_155 MARFEPHPARRPAIVAGASSGIGAATATELAAHGFPVALGARRVEKCQEL
MLBr_00429|M.leprae_Br4923 MPIPAPSPEAR-AVVTGASQNIGEALATELAAHGHSLIVIARREDVLVDL
*. * * * ::::*** .** * * **..*.. : *** : .:
Mb0788c|M.bovis_AF2122/97 VDKIRADGGEAVAF-PLDVTDPESVKSFVAQTVEALGEVELLVSSAGDML
Rv0765c|M.tuberculosis_H37Rv VDKIRADGGEAVAF-PLDVTDPESVKSFVAQTVEALGEVELLVSSAGDML
MAB_1215c|M.abscessus_ATCC_199 VEKIRSDGGEAVAF-PLDVTDADSVKSFVAESIDALGEIDLLVSGAGDMY
Mflv_1597|M.gilvum_PYR-GCK VGKIRADGGEAIAV-HLDVTEPDSVKAAVEQTTSQLGDIEVLVAGAGDTY
Mvan_5160|M.vanbaalenii_PYR-1 VDKIRADGGEAIAV-HLDVTDPDSVKSAVEQTTVELGEIEVLVAGAGDTY
MAV_0709|M.avium_104 VGKINADGGEAVGF-HLDVTDPNSVKSFVAQAVDALGDIEVLVAGAGDTY
MMAR_4931|M.marinum_M VEKIRADGGDAVAL-ALDVTDADSVKDFVHQATERLGDIEVLVAGAGDTY
MUL_0474|M.ulcerans_Agy99 VEKIRADGGDAVAL-ALDVTDADSVKDFVHQATERLGDIEVLVAGAGDTY
TH_1257|M.thermoresistible__bu VEQIRADGGEAVAL-HLDVTDPDSVKSFVHGAAEKLGDIEVLVAGAGDTF
MSMEG_5862|M.smegmatis_MC2_155 VDQITARGGEAVAL-PLDVTDADSVKSFVHAATEALGDIEVLIAGAGDTD
MLBr_00429|M.leprae_Br4923 AARLADKYRVAVEVRPADLTDPSERVKLADELTVRP--ISILCANAGTAT
. :: *: . *:*:... . :.:* :.**
Mb0788c|M.bovis_AF2122/97 PGQLHEVSTEAFAEQVQIHLVGANRLATAVLPAMVARRRGDLIFVGSDVG
Rv0765c|M.tuberculosis_H37Rv PGQLHEVSTEAFAEQVQIHLVGANRLATAVLPAMVARRRGDLIFVGSDVG
MAB_1215c|M.abscessus_ATCC_199 PGRVHEMSAATFAAQIQVHLLGANQLATEVVPDMVSRRRGDVVFIGSDVA
Mflv_1597|M.gilvum_PYR-GCK FGKLDSISTEQFESQIQIHLIGANRVAGAVLPGMIERRRGDLIFVGSDVA
Mvan_5160|M.vanbaalenii_PYR-1 FGKLDAISTEEFESQIQIHLIGANRVATAVLPGMIERRRGDLIFVGSDVA
MAV_0709|M.avium_104 FGKLAEIAGDEFESQLQIHLVGAFRLASAVLPGMLERQRGDLIFVGSDVA
MMAR_4931|M.marinum_M FGRLYEIDTETFESQVQIHLIGANRLATAVLPGMLERQRGDLIFVGSDVA
MUL_0474|M.ulcerans_Agy99 FGRLYEIDTETFESQVQIHLIGANRLATAVLPGMLERQRGDLIFVGSDVA
TH_1257|M.thermoresistible__bu FGRLHEISTEEFESQVQIHLIGANRLATAVLPGMVERRRGDLIFVGSDVA
MSMEG_5862|M.smegmatis_MC2_155 FGRLHEISTEEFEQQVQIHLIGANRMATAVLPGMVERRRGDVIFVGSDVS
MLBr_00429|M.leprae_Br4923 FGPVSALDPAGEKAQVQLNAVAVHDLTLAVLPGMIERKAGGILISGSAAG
* : : *:*:: :.. :: *:* *: *: *.::: ** ..
Mb0788c|M.bovis_AF2122/97 LRQRPHMGAYGAAKAGLAAMVTNLQMELEGTGVRASIVHPGPTLTGMGWQ
Rv0765c|M.tuberculosis_H37Rv LRQRPHMGAYGAAKAGLAAMVTNLQMELEGTGVRASIVHPGPTLTGMGWQ
MAB_1215c|M.abscessus_ATCC_199 LHQRPHMGAYGAAKAALLAMVNTMRMELEGTGVRASIVHPGPTLTSMGWQ
Mflv_1597|M.gilvum_PYR-GCK LRQRPHMGAYGAAKAALVAMVTNYQMELEGTGVRASIVHPGPTKTSMGWS
Mvan_5160|M.vanbaalenii_PYR-1 LRQRPHMGAYGAAKAALVAMVTNYQMELEGTGVRASIVHPGPTKTAMGWS
MAV_0709|M.avium_104 LRQRPHMGAYGAAKAALVAMVNNFQMELEGTGVRASVVHPGPTKTAMGWS
MMAR_4931|M.marinum_M LRQRPHMGAYGAAKAALVAMVTNLQMELEGTGLRASIVHPGPTKTAMGWS
MUL_0474|M.ulcerans_Agy99 LRQRPHMGAYGAAKAALVAMVTNLQMELEGTGLRASIVHPGRTKTAMGWS
TH_1257|M.thermoresistible__bu LRQRPHMGGYGAAKAGLLAMVHNLQMELEGTGVRASIVHPGPTKTAMGWS
MSMEG_5862|M.smegmatis_MC2_155 LRQRPHMGAYGAAKAGLLAMVNNLQMELEGTGVRASIVHPGPTRTSMGWS
MLBr_00429|M.leprae_Br4923 NSPIPYNATYAATKAFANTFSESLRGELHGSGVHVTLLAPGPVRTELPDH
*: . *.*:** :: . : **.*:*::.::: ** . * :
Mb0788c|M.bovis_AF2122/97 LSAEQVGPMLADWAKWGQARHNYFLRPSDLAR-AIAFVAETPRGCVVVNM
Rv0765c|M.tuberculosis_H37Rv LSAEQVGPMLADWAKWGQARHNYFLRPSDLAR-AIAFVAETPRGCVVVNM
MAB_1215c|M.abscessus_ATCC_199 LSAEQVGPMLEDWKSWGQARHSYFLRPSDIAR-AVTFVAETPRGAHIPEM
Mflv_1597|M.gilvum_PYR-GCK LPAELIGPALEDWAKWGQARHDYFLRADDLAR-AITFVAETPRGGFIANM
Mvan_5160|M.vanbaalenii_PYR-1 LPAELIGPALEDWAKWGQARHDYFLRAADLAR-AITFVAETPRGGFIASM
MAV_0709|M.avium_104 LPAEKIGPALEDWAKWGQARHDYFLRAADLAR-AITFVAETPRGGFIANM
MMAR_4931|M.marinum_M LPVESIGPALEDWAKWGQARHDYFLRASDIAR-AITFVAETPRGGFVANM
MUL_0474|M.ulcerans_Agy99 LPVESIGPALEDWAKWGQARHDYFLRASDIAR-AITFVAETPRGGFVANM
TH_1257|M.thermoresistible__bu LPPDRIEQALNDWAKWGQARHDYFLRASDLAR-AIAFVAETPRGGFIANM
MSMEG_5862|M.smegmatis_MC2_155 LPAEKIGPALEDWAKWGQARHDYFLRAADLAR-AITFVAETPRGGFIANL
MLBr_00429|M.leprae_Br4923 DEASLVEKLVPDFL-WISTEHTARLSLDALERNKMRVVPGLTSKAMSVAS
. : : *: * .:.* * : * : .*. .
Mb0788c|M.bovis_AF2122/97 EIQPEAPLRDAPAHRQKLVLGEEGMPG-
Rv0765c|M.tuberculosis_H37Rv EIQPEAPLRDAPAHRQKLVLGEEGMPG-
MAB_1215c|M.abscessus_ATCC_199 EVQPEAPLKDAPADKQKLELGEEGMPS-
Mflv_1597|M.gilvum_PYR-GCK ELQPEAPLSEAKD-RQKLALGEEGMPS-
Mvan_5160|M.vanbaalenii_PYR-1 ELQPEAPLASVKD-RQQLKVDKEALNS-
MAV_0709|M.avium_104 ELQPEAPLADNKD-RQKLALGEEGMPGQ
MMAR_4931|M.marinum_M ELQPEAPLAATTN-RQQLKVDEKAKDS-
MUL_0474|M.ulcerans_Agy99 ELQPEAPLAATTN-RQQLKVDEKAKDS-
TH_1257|M.thermoresistible__bu ELQPEAPLADAPKERQQLTYPETS----
MSMEG_5862|M.smegmatis_MC2_155 EIQPEAPLAEVTD-RQKLALGEEGMPNQ
MLBr_00429|M.leprae_Br4923 RYAPRAIVASIVG-NFYKKLGGG-----
. *.* : .