For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VTYRLTQYAHGGGCACKIPPGELEDVVRGLGAASPRDPLGELLVGLDSGDDAAVVRIQQGTALIATTDFF TPVVDDAYDWGRIAATNALSDVYAMGGRPVVAVNLLGWPREVLPFELAAETLRGGLDVCGLAGCHLAGGH SVDDPEPKYGLAVTGIADPNRLLRNDSGKPGLPLSLTKPLGIGVLNNRHKATGERFEEAIAVMTTLNAEA AGAALAAGVECATDVTGFGLLGHLHKLARASGVTAVLDAAAVPYLDGAREALAAGYLSGGTRRNLDWVAP HVDLSGVGEDEALLLADAQTSGGLLIAGEIPGAPVIGELVPRGEHTIVVR
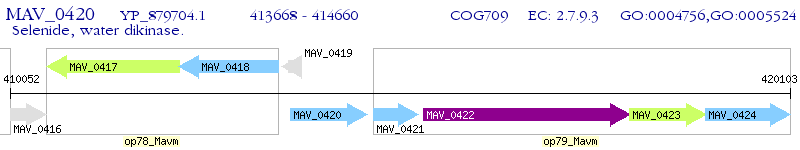
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_0420 | selD | - | 100% (330) | selenide, water dikinase |
| M. avium 104 | MAV_3829 | thiL | 4e-10 | 28.67% (286) | thiamine monophosphate kinase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3001c | thiL | 2e-10 | 30.65% (261) | thiamine monophosphate kinase |
| M. gilvum PYR-GCK | Mflv_4216 | - | 4e-12 | 29.55% (247) | thiamine monophosphate kinase |
| M. tuberculosis H37Rv | Rv2977c | thiL | 2e-10 | 30.65% (261) | thiamine monophosphate kinase |
| M. leprae Br4923 | MLBr_01676 | thiL | 5e-12 | 27.52% (298) | thiamine monophosphate kinase |
| M. abscessus ATCC 19977 | MAB_3284c | - | 3e-06 | 26.44% (295) | thiamine monophosphate kinase |
| M. marinum M | MMAR_5189 | - | 1e-169 | 88.48% (330) | thiamine-monophosphate kinase |
| M. smegmatis MC2 155 | MSMEG_1852 | selD | 1e-169 | 87.88% (330) | selenide, water dikinase |
| M. thermoresistible (build 8) | TH_2051 | thiL | 6e-05 | 24.25% (301) | PROBABLE THIAMINE-MONOPHOSPHATE KINASE THIL |
| M. ulcerans Agy99 | MUL_1978 | thiL | 6e-10 | 28.46% (267) | thiamine monophosphate kinase |
| M. vanbaalenii PYR-1 | Mvan_2151 | - | 3e-10 | 29.50% (278) | thiamine monophosphate kinase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3001c|M.bovis_AF2122/97 ---------MTTKDHSLATESPTLQQLGEFAVIDR--LVRGRRQPA---T
Rv2977c|M.tuberculosis_H37Rv ---------MTTKDHSLATESPTLQQLGEFAVIDR--LVRGRRQPA---T
MLBr_01676|M.leprae_Br4923 ---------MCNHECGALGESVTLEQLGEFAVIDR--LVRGRRQPA---V
MUL_1978|M.ulcerans_Agy99 ------------MGQDTSGQTPTLRELGEFAVIDR--LVQGRNQPA---V
TH_2051|M.thermoresistible__bu ---MAGDQPRDMAGDEAGGRAETLAAVGEFAVIDR--LVAGRRQPG---A
Mflv_4216|M.gilvum_PYR-GCK --------------MAAEDADAKLSELGEFAVIDR--LVADRRQPP---A
Mvan_2151|M.vanbaalenii_PYR-1 --------------MAGEDADDTLSARGEFAVIDR--LTAGRRQPP---F
MAB_3284c|M.abscessus_ATCC_199 --------------------MPNLGDTGEFGVISR--LTAGRDLGA---D
MAV_0420|M.avium_104 -------MTYRLTQYAHGGGCACKIPPGELEDVVRGLGAASPRDPLGELL
MSMEG_1852|M.smegmatis_MC2_155 -------MTYRLTQYAHGGGCACKIPPGELEDVVRGLTHADPKDPLGELL
MMAR_5189|M.marinum_M MGMVTDQGTYRLTQYAHGGGCACKIPPGELEDVVRGLGAAAPRDPVGELL
**: : *
Mb3001c|M.bovis_AF2122/97 VLLGPGDDAALVSAGDGRTVVSTDMLVQDSHFRLDWSTPQDVGRKAIAQN
Rv2977c|M.tuberculosis_H37Rv VLLGPGDDAALVSAGDGRTVVSTDMLVQDSHFRLDWSTPQDVGRKAIAQN
MLBr_01676|M.leprae_Br4923 VALGPGDDAAVVTASDGRIVVSTDVLVQDRHFRLDWSTPHDVGRKAIAQN
MUL_1978|M.ulcerans_Agy99 VELGPGDDAAVVAAPDGRTVISTDMLVQDRHFRLDWSSPHDVGRKAIAQN
TH_2051|M.thermoresistible__bu VALGPGDDAAVVHAADGRTVVCTDMLVQDRHFRLDWSTPHQVGRKAIAQN
Mflv_4216|M.gilvum_PYR-GCK VSLGPGDDAAVVFAADGRTVVCTDMLVEGRHFRLDWSTPHDIGRKAIAQN
Mvan_2151|M.vanbaalenii_PYR-1 VTLGPGDDAAVVLAPDGRVVVCTDMLVENRHFRLDWSSPHDVGRKAIAQN
MAB_3284c|M.abscessus_ATCC_199 VLLGPGDDAAVVTVPDGRVVVSTDMLVQDRHFRLEWSSPSDIGRKAVAQN
MAV_0420|M.avium_104 VGLDSGDDAAVVRIQQG-----TALIATTDFFTPVVDDAYDWGRIAATNA
MSMEG_1852|M.smegmatis_MC2_155 VGLDDGDDAAAVRINGD-----TALIATTDFFTPVVDDAYDWGRIAATNA
MMAR_5189|M.marinum_M VGLESGDDAAAVLIAEG-----TALIATTDFFTPVVDDPYDWGRIAATNA
* * ***** * . * ::. .* . . : ** * ::
Mb3001c|M.bovis_AF2122/97 AADIEAMGARATAFVVGFGAP-AETPAAQASALVDGMWEEAGRIGAGIVG
Rv2977c|M.tuberculosis_H37Rv AADIEAMGARATAFVVGFGAP-AETPAAQASALVDGMWEEAGRIGAGIVG
MLBr_01676|M.leprae_Br4923 AADIEAMGARATTFVVGLGAP-GDTPVAQVDALVDGLWDEAGRIGAGIVG
MUL_1978|M.ulcerans_Agy99 AADIAAMGARPTAFVVSLGAP-AHTPVAEVTALLDGMWEEAGRIGTGIVG
TH_2051|M.thermoresistible__bu AADIAAMGARSTAFVVAFGAP-ADTPAAAALALADGLWAEAQSLGAGIVG
Mflv_4216|M.gilvum_PYR-GCK AADIEAMGARATAFVVAFGAP-GDTPTAAAVQVADGMWHEAALLGAGIVG
Mvan_2151|M.vanbaalenii_PYR-1 AADIEAMGGRSTAFVVAFGAP-GDTDTASAVELADGMWHEAGLMGAGIVG
MAB_3284c|M.abscessus_ATCC_199 AADIEAMGGRVTAFVVAVGAP-AQTDIAFLDQLNEGVWAEAALVQAPIVG
MAV_0420|M.avium_104 LSDVYAMGGRPVVAVNLLGWPREVLPFELAAETLRGGLDVCGLAGCHLAG
MSMEG_1852|M.smegmatis_MC2_155 LSDVYAMGGRPVVAVNLLGWPRDVLPFELAAETLRGGLDVCAAAGCHLAG
MMAR_5189|M.marinum_M LSDVYAMGGRPVVAVNLLGWPRDVLPFELATETLRGGRDVCASAGCHLAG
:*: ***.* .. * .* * * . :.*
Mb3001c|M.bovis_AF2122/97 GDLVSCRQWVVSVTAIGDLDGRAPVLRSGAKAGSVLAVVGELGRSAAGYA
Rv2977c|M.tuberculosis_H37Rv GDLVSCRQWVVSVTAIGDLDGRAPVLRSGAKAGSVLAVVGELGRSAAGYA
MLBr_01676|M.leprae_Br4923 GDLVSCPQWVLSVTVLGDLDGRAPVLRSGAKNGSMVAVAGALGRSAAGYA
MUL_1978|M.ulcerans_Agy99 GDMVRCPQWVVSVTVLGDLEGRPPVTRGGAKPGSSVAVAGELGRSAAGYA
TH_2051|M.thermoresistible__bu GDLVRAPNWVVSVTALGDLEGRPPVPRGGARAGDVVAAVGQLGRSAAGYA
Mflv_4216|M.gilvum_PYR-GCK GDTVAAPQWVISVAALGDLGGREPVRRDGARPGDTVAVTGVLGRSAAGYE
Mvan_2151|M.vanbaalenii_PYR-1 GDVVAAPQWVVSVAALGDLGGRDPVRRDGAKPGDMVAVVGELGRSQAGFT
MAB_3284c|M.abscessus_ATCC_199 GDLVSARELVVSVTVLGDLQGRAPVTRSGAVVGDTVAICGALGTSAAGYR
MAV_0420|M.avium_104 GHSVDDPEPKYGLAVTGIADPNRLLRNDSGKPGLPLSLTKPLG---IGVL
MSMEG_1852|M.smegmatis_MC2_155 GHSVDDPEPKYGLAVTGIADPKRLLRNDSGKPGVPLSLTKPLG---IGVL
MMAR_5189|M.marinum_M GHSVDDPEPKYGLAVTGIADPNRLLRNDSGRPGTPLSLTKPLG---IGVL
*. * : .::. * . : .... * :: ** *
Mb3001c|M.bovis_AF2122/97 LWCNGIEDFAELRRRHLVPQPPYGHGAAAAAVGAQAMIDVS-DGLLADLR
Rv2977c|M.tuberculosis_H37Rv LWCNGIEDFAELRRRHLVPQPPYGHGAAAAAVGAQAMIDVS-DGLLADLR
MLBr_01676|M.leprae_Br4923 LWHKGIDGFDDLRRRHLVPEPPYGKGVAAAACGAQAMIDVS-DGLIADLR
MUL_1978|M.ulcerans_Agy99 LWHNEIAGFDELRRRHLVPDPPYGQGVAAAVAGAQAMIDVS-DGLVADLR
TH_2051|M.thermoresistible__bu LWRNGIKDFPALRERHLVPAPPYRQGPVAASSGATSMIDVS-DGLVADLG
Mflv_4216|M.gilvum_PYR-GCK LLRNGIDGFDELRRQHRVPSPPYGQGAIAAQAGATAMTDVS-DGLLADLA
Mvan_2151|M.vanbaalenii_PYR-1 LWDSGISGHDALRRRHLVPEPPYGQGEIAARGGATAMTDVS-DGLLADLG
MAB_3284c|M.abscessus_ATCC_199 LWQEQIDEFPDLRRVHLVPAPPYGQGPAAARAGATAMTDVS-DGLLADLA
MAV_0420|M.avium_104 NNRHKATGERFEEAIAVMTTLNAEAAGAALAAGVECATDVTGFGLLGHLH
MSMEG_1852|M.smegmatis_MC2_155 NSRHKATGEVFPQAIEAMTTLNAAASAAALAAGVECATDVTGFGLLGHLH
MMAR_5189|M.marinum_M NSRHKATGERFAQAIDAMTTLNADAAAAAVAAGVECATDITGFGLLGHLH
. :. . * *. . *:: **:..*
Mb3001c|M.bovis_AF2122/97 HIAEASGVRIDLS---AAALAADRDALTAAATALGTDPWPWVLSGGEDHA
Rv2977c|M.tuberculosis_H37Rv HIAEASGVRIDLS---AAALAADRDALTAAATALGTDPWPWVLSGGEDHA
MLBr_01676|M.leprae_Br4923 HVARASGVSIDLS---TAALAADHAALIAAATAVGGDPWPWVLGGGEDHA
MUL_1978|M.ulcerans_Agy99 HIADAAAVGIDVA---TAALAQDHRALIAAADAVGADPCGWVLGGGEDHA
TH_2051|M.thermoresistible__bu HVAESSAVGIDVF---TDALAGDHAAVAPAAEVTGADAWSWVLGGGEDHA
Mflv_4216|M.gilvum_PYR-GCK HIASASGVSIDIT---LDTLTSDEAAVADAAGATGVDARQWVLGGGEDHA
Mvan_2151|M.vanbaalenii_PYR-1 HIASASEVSIDLA---IGHLQPDIDALAEAAAAVGADARQWVLGGGEDHA
MAB_3284c|M.abscessus_ATCC_199 HIAELSAVHIDLS---SPSLEPYLQPVAGAAGLLGADPWEWVLTGGEDHA
MAV_0420|M.avium_104 KLARASGVTAVLDAAAVPYLDGAREALAAGYLSGGTRRNLDWVAPHVDLS
MSMEG_1852|M.smegmatis_MC2_155 KLARASGVTAVIDSKAVPYLEGAREAIAAGFVSGGSRRNLEWVSPHIDVS
MMAR_5189|M.marinum_M KLARASGVTAVIDSTAVPYLDGARQALAAGYVSGGTRRNLEWVGPHVDLS
::* : * : * .: . * : * :
Mb3001c|M.bovis_AF2122/97 LVACFVGPVPAGWRTIGRVL----DGPARVLVDGEEWTGYAGWQSFGEPD
Rv2977c|M.tuberculosis_H37Rv LVACFVGPVPAGWRTIGRVL----DGPARVLVDGEEWTGYAGWQSFGEPD
MLBr_01676|M.leprae_Br4923 LVACFAGTALSGWRIIGRVF----DGPARVLVDGQEWRGHAGWQSFGG--
MUL_1978|M.ulcerans_Agy99 LVACFSGAVPDGWRVIGRVL----DGPARVLVDGQQWSGYAGWQSFD---
TH_2051|M.thermoresistible__bu LVATFPGEPPPGWRVIGRVV----DGPARVLVDGVAWSGSAGWESFGD--
Mflv_4216|M.gilvum_PYR-GCK LVATFPGAVPPNWRVLGTVAATAADGAPAVTVDGSPWIGNAGWQSF----
Mvan_2151|M.vanbaalenii_PYR-1 LVATFASQPPPGWRVIGTVG--ASDGTTAVTVDGARWRGSAGWQSF----
MAB_3284c|M.abscessus_ATCC_199 LVGMFPGVVPTGWVPIGRVT----VGEPGVTVDGAAWTRATGWDSFRLS-
MAV_0420|M.avium_104 GVGEDEALLLADAQTSGGLLIAG--EIPGAPVIGELVPR-GEHTIVVR--
MSMEG_1852|M.smegmatis_MC2_155 EVKEDEALLLADAQTSGGLLIAG--EIPGAPVIGELVAR-GDHTIVVR--
MMAR_5189|M.marinum_M AVDESEALLLADAQTSGGLLIAG--EIPGAPVIGELVARRAGHTIVVR--
* . . * : . . * * .
Mb3001c|M.bovis_AF2122/97 NQGSLG
Rv2977c|M.tuberculosis_H37Rv NQGSLG
MLBr_01676|M.leprae_Br4923 ------
MUL_1978|M.ulcerans_Agy99 ------
TH_2051|M.thermoresistible__bu ------
Mflv_4216|M.gilvum_PYR-GCK ------
Mvan_2151|M.vanbaalenii_PYR-1 ------
MAB_3284c|M.abscessus_ATCC_199 ------
MAV_0420|M.avium_104 ------
MSMEG_1852|M.smegmatis_MC2_155 ------
MMAR_5189|M.marinum_M ------