For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
AAEDADAKLSELGEFAVIDRLVADRRQPPAVSLGPGDDAAVVFAADGRTVVCTDMLVEGRHFRLDWSTPH DIGRKAIAQNAADIEAMGARATAFVVAFGAPGDTPTAAAVQVADGMWHEAALLGAGIVGGDTVAAPQWVI SVAALGDLGGREPVRRDGARPGDTVAVTGVLGRSAAGYELLRNGIDGFDELRRQHRVPSPPYGQGAIAAQ AGATAMTDVSDGLLADLAHIASASGVSIDITLDTLTSDEAAVADAAGATGVDARQWVLGGGEDHALVATF PGAVPPNWRVLGTVAATAADGAPAVTVDGSPWIGNAGWQSF*
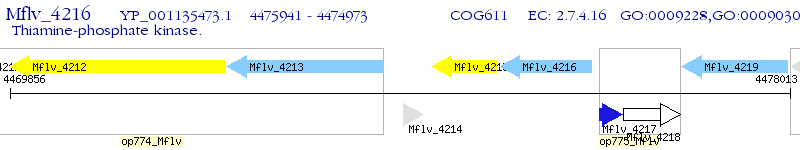
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_4216 | - | - | 100% (322) | thiamine monophosphate kinase |
| M. gilvum PYR-GCK | Mflv_3982 | - | 2e-08 | 27.56% (254) | hydrogenase expression/formation protein HypE |
| M. gilvum PYR-GCK | Mflv_1636 | - | 5e-05 | 23.21% (336) | phosphoribosylaminoimidazole synthetase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3001c | thiL | 1e-114 | 66.45% (313) | thiamine monophosphate kinase |
| M. tuberculosis H37Rv | Rv2977c | thiL | 1e-114 | 66.45% (313) | thiamine monophosphate kinase |
| M. leprae Br4923 | MLBr_01676 | thiL | 1e-116 | 66.45% (313) | thiamine monophosphate kinase |
| M. abscessus ATCC 19977 | MAB_3284c | - | 1e-100 | 61.34% (313) | thiamine monophosphate kinase |
| M. marinum M | MMAR_1735 | thiL | 1e-115 | 66.45% (313) | thiamine-monophosphate kinase ThiL |
| M. avium 104 | MAV_3829 | thiL | 1e-117 | 66.77% (316) | thiamine monophosphate kinase |
| M. smegmatis MC2 155 | MSMEG_2398 | thiL | 1e-119 | 68.12% (320) | thiamine monophosphate kinase |
| M. thermoresistible (build 8) | TH_2051 | thiL | 1e-125 | 68.92% (325) | PROBABLE THIAMINE-MONOPHOSPHATE KINASE THIL |
| M. ulcerans Agy99 | MUL_1978 | thiL | 1e-115 | 66.77% (313) | thiamine monophosphate kinase |
| M. vanbaalenii PYR-1 | Mvan_2151 | - | 1e-139 | 76.09% (322) | thiamine monophosphate kinase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_4216|M.gilvum_PYR-GCK -------------MAAEDAD---AKLSELGEFAVIDRLVADRRQPPAVSL
Mvan_2151|M.vanbaalenii_PYR-1 -------------MAGEDAD---DTLSARGEFAVIDRLTAGRRQPPFVTL
Mb3001c|M.bovis_AF2122/97 ------MTTKDHSLATESP-----TLQQLGEFAVIDRLVRGRRQPATVLL
Rv2977c|M.tuberculosis_H37Rv ------MTTKDHSLATESP-----TLQQLGEFAVIDRLVRGRRQPATVLL
MLBr_01676|M.leprae_Br4923 ------MCNHECGALGESV-----TLEQLGEFAVIDRLVRGRRQPAVVAL
MAV_3829|M.avium_104 --------------MRDES-----TLRQLGEFGVIDRLVAGRRQPAAVTL
MMAR_1735|M.marinum_M ---------MGQDTSGQTP-----TLRELGEFAVIDRLVQGRNQPAVVEL
MUL_1978|M.ulcerans_Agy99 ---------MGQDTSGQTP-----TLRELGEFAVIDRLVQGRNQPAVVEL
TH_2051|M.thermoresistible__bu -----MAGDQPRDMAGDEAGGRAETLAAVGEFAVIDRLVAGRRQPGAVAL
MSMEG_2398|M.smegmatis_MC2_155 MAEDTAGANDAQTSPGPFAGSASDSLSAVGEFAVIDRLLNGRVQPPSVTV
MAB_3284c|M.abscessus_ATCC_199 -------------MP---------NLGDTGEFGVISRLTAGRDLGADVLL
.* ***.**.** .* * :
Mflv_4216|M.gilvum_PYR-GCK GPGDDAAVVFAADGRTVVCTDMLVEGRHFRLDWSTPHDIGRKAIAQNAAD
Mvan_2151|M.vanbaalenii_PYR-1 GPGDDAAVVLAPDGRVVVCTDMLVENRHFRLDWSSPHDVGRKAIAQNAAD
Mb3001c|M.bovis_AF2122/97 GPGDDAALVSAGDGRTVVSTDMLVQDSHFRLDWSTPQDVGRKAIAQNAAD
Rv2977c|M.tuberculosis_H37Rv GPGDDAALVSAGDGRTVVSTDMLVQDSHFRLDWSTPQDVGRKAIAQNAAD
MLBr_01676|M.leprae_Br4923 GPGDDAAVVTASDGRIVVSTDVLVQDRHFRLDWSTPHDVGRKAIAQNAAD
MAV_3829|M.avium_104 GPGDDAALVTTGDGRVVVSTDMLVQDRHFRLDWSTPHDIGRKAIAQNAAD
MMAR_1735|M.marinum_M GPGDDAAVVAAPDGRTVISTDMLVQDRHFRLDWSSPHDVGRKAIAQNAAD
MUL_1978|M.ulcerans_Agy99 GPGDDAAVVAAPDGRTVISTDMLVQDRHFRLDWSSPHDVGRKAIAQNAAD
TH_2051|M.thermoresistible__bu GPGDDAAVVHAADGRTVVCTDMLVQDRHFRLDWSTPHQVGRKAIAQNAAD
MSMEG_2398|M.smegmatis_MC2_155 GPGDDAAVIATSDGRTAVSTDMLVERRHFRLDWASPHQIGRKAIAQNAAD
MAB_3284c|M.abscessus_ATCC_199 GPGDDAAVVTVPDGRVVVSTDMLVQDRHFRLEWSSPSDIGRKAVAQNAAD
*******:: . *** .:.**:**: ****:*::* ::****:******
Mflv_4216|M.gilvum_PYR-GCK IEAMGARATAFVVAFGAPGDTPTAAAVQVADGMWHEAALLGAGIVGGDTV
Mvan_2151|M.vanbaalenii_PYR-1 IEAMGGRSTAFVVAFGAPGDTDTASAVELADGMWHEAGLMGAGIVGGDVV
Mb3001c|M.bovis_AF2122/97 IEAMGARATAFVVGFGAPAETPAAQASALVDGMWEEAGRIGAGIVGGDLV
Rv2977c|M.tuberculosis_H37Rv IEAMGARATAFVVGFGAPAETPAAQASALVDGMWEEAGRIGAGIVGGDLV
MLBr_01676|M.leprae_Br4923 IEAMGARATTFVVGLGAPGDTPVAQVDALVDGLWDEAGRIGAGIVGGDLV
MAV_3829|M.avium_104 IEAMGARPTAFVVGFGAPGQTAAAQVDALVDGMWDEAQRVGAGIAGGDLV
MMAR_1735|M.marinum_M IAAMGARPTAFVVSLGAPAHTPVAEVIALLDGMWEEAGRIGTGIVGGDMV
MUL_1978|M.ulcerans_Agy99 IAAMGARPTAFVVSLGAPAHTPVAEVTALLDGMWEEAGRIGTGIVGGDMV
TH_2051|M.thermoresistible__bu IAAMGARSTAFVVAFGAPADTPAAAALALADGLWAEAQSLGAGIVGGDLV
MSMEG_2398|M.smegmatis_MC2_155 IEAMGARPAAFVVSFGAPSDTPADAAVELSDGMWDEARRCGGGIAGGDLV
MAB_3284c|M.abscessus_ATCC_199 IEAMGGRVTAFVVAVGAPAQTDIAFLDQLNEGVWAEAALVQAPIVGGDLV
* ***.* ::***..***..* : :*:* ** *.*** *
Mflv_4216|M.gilvum_PYR-GCK AAPQWVISVAALGDLGGREPVRRDGARPGDTVAVTGVLGRSAAGYELLRN
Mvan_2151|M.vanbaalenii_PYR-1 AAPQWVVSVAALGDLGGRDPVRRDGAKPGDMVAVVGELGRSQAGFTLWDS
Mb3001c|M.bovis_AF2122/97 SCRQWVVSVTAIGDLDGRAPVLRSGAKAGSVLAVVGELGRSAAGYALWCN
Rv2977c|M.tuberculosis_H37Rv SCRQWVVSVTAIGDLDGRAPVLRSGAKAGSVLAVVGELGRSAAGYALWCN
MLBr_01676|M.leprae_Br4923 SCPQWVLSVTVLGDLDGRAPVLRSGAKNGSMVAVAGALGRSAAGYALWHK
MAV_3829|M.avium_104 SCPQWVISVTALGELDGRSPVCRAGAEPGALIAVAGDLGRSAAGFDLWRN
MMAR_1735|M.marinum_M RCPQWVVSVTVLGDLEGRPPVTRGGAKPGSSVAVAGELGRSAAGYALWHN
MUL_1978|M.ulcerans_Agy99 RCPQWVVSVTVLGDLEGRPPVTRGGAKPGSSVAVAGELGRSAAGYALWHN
TH_2051|M.thermoresistible__bu RAPNWVVSVTALGDLEGRPPVPRGGARAGDVVAAVGQLGRSAAGYALWRN
MSMEG_2398|M.smegmatis_MC2_155 RAPLWVISVTVLGDLGGRAPVLRSGARAGDVVAVAGDLGRSAAGYALWAK
MAB_3284c|M.abscessus_ATCC_199 SARELVVSVTVLGDLQGRAPVTRSGAVVGDTVAICGALGTSAAGYRLWQE
. *:**:.:*:* ** ** * ** * :* * ** * **: * .
Mflv_4216|M.gilvum_PYR-GCK GIDGFDELRRQHRVPSPPYGQGAIAAQAGATAMTDVSDGLLADLAHIASA
Mvan_2151|M.vanbaalenii_PYR-1 GISGHDALRRRHLVPEPPYGQGEIAARGGATAMTDVSDGLLADLGHIASA
Mb3001c|M.bovis_AF2122/97 GIEDFAELRRRHLVPQPPYGHGAAAAAVGAQAMIDVSDGLLADLRHIAEA
Rv2977c|M.tuberculosis_H37Rv GIEDFAELRRRHLVPQPPYGHGAAAAAVGAQAMIDVSDGLLADLRHIAEA
MLBr_01676|M.leprae_Br4923 GIDGFDDLRRRHLVPEPPYGKGVAAAACGAQAMIDVSDGLIADLRHVARA
MAV_3829|M.avium_104 QIGGFDELRRRHAVPQPPYGQGAVAAAGGAQAMIDVSDGLIADLRHVAEA
MMAR_1735|M.marinum_M EIAGFDELRRRHLVPDPPYGQGVAAAVAGAQAMIDVSDGLVADLRHIADS
MUL_1978|M.ulcerans_Agy99 EIAGFDELRRRHLVPDPPYGQGVAAAVAGAQAMIDVSDGLVADLRHIADA
TH_2051|M.thermoresistible__bu GIKDFPALRERHLVPAPPYRQGPVAASSGATSMIDVSDGLVADLGHVAES
MSMEG_2398|M.smegmatis_MC2_155 GIDEYPELRERHLVPRPPYGHGVAAADAGATAMTDVSDGLIADLGHIATA
MAB_3284c|M.abscessus_ATCC_199 QIDEFPDLRRVHLVPAPPYGQGPAAARAGATAMTDVSDGLLADLAHIAEL
* . **. * ** *** :* ** ** :* ******:*** *:*
Mflv_4216|M.gilvum_PYR-GCK SGVSIDITLDTLTSDEAAVADAAGATGVDARQWVLGGGEDHALVATFPGA
Mvan_2151|M.vanbaalenii_PYR-1 SEVSIDLAIGHLQPDIDALAEAAAAVGADARQWVLGGGEDHALVATFASQ
Mb3001c|M.bovis_AF2122/97 SGVRIDLSAAALAADRDALTAAATALGTDPWPWVLSGGEDHALVACFVGP
Rv2977c|M.tuberculosis_H37Rv SGVRIDLSAAALAADRDALTAAATALGTDPWPWVLSGGEDHALVACFVGP
MLBr_01676|M.leprae_Br4923 SGVSIDLSTAALAADHAALIAAATAVGGDPWPWVLGGGEDHALVACFAGT
MAV_3829|M.avium_104 SGVGMDVSTAALAADRDAVAAAAAAVGVDAWPWVLTGGEDHALVACFPGP
MMAR_1735|M.marinum_M AAVGIDVATAALAQDHRALIAAADAVGADPWGWVLGGGEDHALVACFSGA
MUL_1978|M.ulcerans_Agy99 AAVGIDVATAALAQDHRALIAAADAVGADPCGWVLGGGEDHALVACFSGA
TH_2051|M.thermoresistible__bu SAVGIDVFTDALAGDHAAVAPAAEVTGADAWSWVLGGGEDHALVATFPGE
MSMEG_2398|M.smegmatis_MC2_155 SGVEIDIATDLLASDHAAVAAAAATVAADAWSLVLGGGEDHALVATFPGE
MAB_3284c|M.abscessus_ATCC_199 SAVHIDLSSPSLEPYLQPVAGAAGLLGADPWEWVLTGGEDHALVGMFPGV
: * :*: * .: ** . *. ** ********. * .
Mflv_4216|M.gilvum_PYR-GCK VPPNWRVLGTVAATAADGAPAVTVDGSPWIGNAGWQSF----------
Mvan_2151|M.vanbaalenii_PYR-1 PPPGWRVIGTVG--ASDGTTAVTVDGARWRGSAGWQSF----------
Mb3001c|M.bovis_AF2122/97 VPAGWRTIGRVLDG--PAR--VLVDGEEWTGYAGWQSFGEPDNQGSLG
Rv2977c|M.tuberculosis_H37Rv VPAGWRTIGRVLDG--PAR--VLVDGEEWTGYAGWQSFGEPDNQGSLG
MLBr_01676|M.leprae_Br4923 ALSGWRIIGRVFDG--PAR--VLVDGQEWRGHAGWQSFGG--------
MAV_3829|M.avium_104 APAGWRIIGRVLDG--PAR--VLVDEREWSGPAGWQSY----------
MMAR_1735|M.marinum_M VPDGWRVIGRVLDG--PAR--VLVDGQQWSGYAGWQSFD---------
MUL_1978|M.ulcerans_Agy99 VPDGWRVIGRVLDG--PAR--VLVDGQQWSGYAGWQSFD---------
TH_2051|M.thermoresistible__bu PPPGWRVIGRVVDG--PAR--VLVDGVAWSGSAGWESFGD--------
MSMEG_2398|M.smegmatis_MC2_155 VPDGWRVIGRVRASHAGAR--VLVDGEKWSGTAGWQSFD---------
MAB_3284c|M.abscessus_ATCC_199 VPTGWVPIGRVTVG--EPG--VTVDGAAWTRATGWDSFRLS-------
.* :* * * ** * :**:*: