For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
THPIRIGVQLQPQHAPQYNQIRDAVRRCEDIGVDIAFNWDHFFPLYGDPDGAHFECWTMLGAWAEQTSRI QIGALVTCNSYRNPELLADMARTVDHISEGRLILGIGSGWKQKDYDEYGYEFGTAGSRLDDLAGALPRIE ARLAKLNPAPTRDIPVLIGGGGERKTLRLVAQHADIWHSFTSAEEYPAKSEVLARHCADVGRDPDRIERS AAVKGGAELLANAEALVGLGVTLLTVGCDGPDYDLRDAQALCKWRDGR*
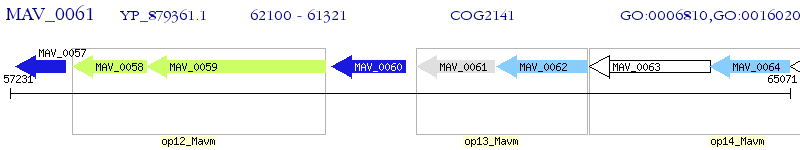
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_0061 | - | - | 100% (259) | MmcI protein |
| M. avium 104 | MAV_2865 | - | 7e-38 | 42.36% (203) | hypothetical protein MAV_2865 |
| M. avium 104 | MAV_0169 | - | 3e-33 | 35.29% (255) | hypothetical protein MAV_0169 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0045c | - | 1e-128 | 83.98% (256) | oxidoreductase |
| M. gilvum PYR-GCK | Mflv_0860 | - | 1e-129 | 82.17% (258) | hypothetical protein Mflv_0860 |
| M. tuberculosis H37Rv | Rv0044c | - | 1e-128 | 83.98% (256) | oxidoreductase |
| M. leprae Br4923 | MLBr_00131 | - | 3e-10 | 29.11% (213) | putative oxidoreductase |
| M. abscessus ATCC 19977 | MAB_3016c | - | 2e-69 | 50.91% (275) | hypothetical protein MAB_3016c |
| M. marinum M | MMAR_0063 | - | 1e-134 | 86.72% (256) | oxidoreductase |
| M. smegmatis MC2 155 | MSMEG_6907 | - | 1e-120 | 80.71% (254) | MmcI protein |
| M. thermoresistible (build 8) | TH_0517 | - | 1e-116 | 77.95% (254) | MmcI protein |
| M. ulcerans Agy99 | MUL_0062 | - | 1e-133 | 86.33% (256) | oxidoreductase |
| M. vanbaalenii PYR-1 | Mvan_6045 | - | 1e-125 | 78.54% (261) | hypothetical protein Mvan_6045 |
CLUSTAL 2.0.9 multiple sequence alignment
Mb0045c|M.bovis_AF2122/97 MTSLVRPDLPVRIGVQLQPQHAPHYRAVRDAVRRCEDIGVDIAFTWDHFF
Rv0044c|M.tuberculosis_H37Rv MTSLVRPDLPVRIGVQLQPQHAPHYRAVRDAVRRCEDIGVDIAFTWDHFF
MMAR_0063|M.marinum_M MTSPVHPDRPLRIGVQLQPQHASHYGDIRDAVRRCEDIGVDIAFNWDHFF
MUL_0062|M.ulcerans_Agy99 MTSPVHPDRPLRIGVQLQPQHASHYGDIRDAVRRCEDIGVDIAFNWDHFF
MAV_0061|M.avium_104 MT------HPIRIGVQLQPQHAPQYNQIRDAVRRCEDIGVDIAFNWDHFF
Mflv_0860|M.gilvum_PYR-GCK --MTSETSRPIRIGVQLQPQHSPRYDLMRDAVRRCEDIGVDVVFNWDHFF
Mvan_6045|M.vanbaalenii_PYR-1 ------MTFPIRIGVQLQPQHSPRYGHLRDAVRRCEDLGVDVAFNWDHFF
MSMEG_6907|M.smegmatis_MC2_155 --------------MQLQPQHSPEYRFIRDAVRRCEDIGVDIAFNWDHFF
TH_0517|M.thermoresistible__bu --------------VQLQPQHSPDYGNLRDAVRRAEDIGVDIAFNWDHFF
MAB_3016c|M.abscessus_ATCC_199 ------MTDRIRIGIQLRPAKTQSYKQWRDAVLRAEDKGADVIFGYDHFH
MLBr_00131|M.leprae_Br4923 -----MAGFRFGFVDALVHTLFPPSLPARASIVSGAVLGADSYWVGDHLN
* * :: *.* : **:
Mb0045c|M.bovis_AF2122/97 PLYG----------------DPDGPHFECWTVLG--AWAEQTSHIEIGAL
Rv0044c|M.tuberculosis_H37Rv PLYG----------------DPDGPHFECWTVLG--AWAEQTSHIEIGAL
MMAR_0063|M.marinum_M PLFG----------------DPDGAHFECWTMLA--AWAEQTSRIQIGAL
MUL_0062|M.ulcerans_Agy99 PLFG----------------DPDGAHFECWTMLA--AWAEQTSRIQIGAL
MAV_0061|M.avium_104 PLYG----------------DPDGAHFECWTMLG--AWAEQTSRIQIGAL
Mflv_0860|M.gilvum_PYR-GCK PLYG----------------DPDGAHFECWTMLG--AWAEQTSRVEIGAL
Mvan_6045|M.vanbaalenii_PYR-1 PLYG----------------DPDGAHFECWTMLG--AWAEQTSRIEIGAL
MSMEG_6907|M.smegmatis_MC2_155 PLYG----------------DPDGAHYECWTMLG--AWAEQTSRIEIGAL
TH_0517|M.thermoresistible__bu PLYG----------------DPDGAHFECWTMLG--AWAEQTSRIEIGAL
MAB_3016c|M.abscessus_ATCC_199 WPAGTLSSQGVVL----DEVQPDVNNFEGWTALG--SWAEVTSRAEIGLL
MLBr_00131|M.leprae_Br4923 ALVPRSVATPKYLGVAAKVVPKIDANYEPWTMLGNLAAGNRLNGLRLGVC
::* ** *. : .: . .:*
Mb0045c|M.bovis_AF2122/97 VTCNSYRNPELLADMARTVDHISGGRLILGIGSGWKQKDYDEYGYRFGTA
Rv0044c|M.tuberculosis_H37Rv VTCNSYRNPELLADMARTVDHISGGRLILGIGSGWKQKDYDEYGYRFGTA
MMAR_0063|M.marinum_M VTCNSYRNPELLADMARTVDHICDGRLILGIGSGWKQKDYDEYGYEFGTA
MUL_0062|M.ulcerans_Agy99 VTCNSYRNPELLADMARTVDHICDGRLILGIGSGWKQKDYDEYGYEFGTA
MAV_0061|M.avium_104 VTCNSYRNPELLADMARTVDHISEGRLILGIGSGWKQKDYDEYGYEFGTA
Mflv_0860|M.gilvum_PYR-GCK VTCNSYRNPELLADMARTVDHISDGRLILGIGSGWKQKDYDEYGYEFGTV
Mvan_6045|M.vanbaalenii_PYR-1 VSCNSYRNPELLADMARTVDHISDGRLILGIGSGWKRKDYDEYGYEFGTA
MSMEG_6907|M.smegmatis_MC2_155 VTCNSYRNPELLADMARTVDHISDGRLILGIGSGWKQKDYDEYGYEFGTA
TH_0517|M.thermoresistible__bu VTCNSYRNPDLLADMARTVDHISGGRLIFGIGSGWFRRDYDEYGYEFGTA
MAB_3016c|M.abscessus_ATCC_199 VTGVGYRNPDLLADMARTVDHISGGRLILGVGAGWYERDYTTYGYDFGTT
MLBr_00131|M.leprae_Br4923 VTDAGRRNPAVTAQAAATLHLLTRGKAMLGIGVGERE-GNEPYGVEWTKP
*: . *** : *: * *:. : *: ::*:* * . . ** : .
Mb0045c|M.bovis_AF2122/97 GSRLDDLAAALPRIKARLGKLN----------------PPPTRDIPVLIG
Rv0044c|M.tuberculosis_H37Rv GSRLDDLAAALPRIKARLGKLN----------------PPPTRDIPVLIG
MMAR_0063|M.marinum_M GSRLDDLAGALPRIKTRLAKLN----------------PAPTRDIPVLIG
MUL_0062|M.ulcerans_Agy99 GSRLDDLAGALPRIKTRLAKLI----------------PAPTRDIPVLIG
MAV_0061|M.avium_104 GSRLDDLAGALPRIEARLAKLN----------------PAPTRDIPVLIG
Mflv_0860|M.gilvum_PYR-GCK GSRLDDLAAALPRIESRLSKLN----------------PQPVRDIPVLIG
Mvan_6045|M.vanbaalenii_PYR-1 GSRLDDLAVAMPRIEARLDKLN----------------PQPVRDIPVLIG
MSMEG_6907|M.smegmatis_MC2_155 GSRLDDLAAALPRIRARLDKLN----------------PAPTREIPILIG
TH_0517|M.thermoresistible__bu GGRLDDLAAALPRIKARWAKLN----------------PPPTRELPILIG
MAB_3016c|M.abscessus_ATCC_199 GSRFDLFEDSLVRIESRLGQLL----------------PQPVRPIPILIG
MLBr_00131|M.leprae_Br4923 VARFQEALATIRALWDSNGELVSRESQFFPLHNALFDLPPYRGKWPEIWV
.*:: :: : :* * * :
Mb0045c|M.bovis_AF2122/97 GGGERKTLRLVAEYADIWH--SFTAGDSYLAKSAVLSTHCSTVGRNPATI
Rv0044c|M.tuberculosis_H37Rv GGGERKTLRLVAEYADIWH--SFTAGDSYLAKSAVLSTHCSTVGRNPATI
MMAR_0063|M.marinum_M GGGERKTLRLVAEHADIWH--SFTGVDEFPAKSAVLARHCADVGRDPATI
MUL_0062|M.ulcerans_Agy99 GGGERKTLRLVAEHADIWH--SFTGVDEFPAKSAVLARHCADVGRDPATI
MAV_0061|M.avium_104 GGGERKTLRLVAQHADIWH--SFTSAEEYPAKSEVLARHCADVGRDPDRI
Mflv_0860|M.gilvum_PYR-GCK GGGERKTLRMVAQHAHIWH--SFSDTETYPRKSEILARHCADVGRDPDAI
Mvan_6045|M.vanbaalenii_PYR-1 GGGEKKTLRLVARHAQIWH--SFSDTDSYPRKSDILARHCAEAGRDPATI
MSMEG_6907|M.smegmatis_MC2_155 GQGERKTLPLVAEHAHIWH--AFVDRNTYPGKAEVLDSHCESVGRDAASI
TH_0517|M.thermoresistible__bu GMGEKKTLRLVAEHAHIWH--GFVDRSTYPHKSRVLAEHCAAVGRDPDTI
MAB_3016c|M.abscessus_ATCC_199 GGGEKRTIPLVARHAHIWH--AFGDIDTYRRKNSVLIAEADRIGRDHNEI
MLBr_00131|M.leprae_Br4923 AAHGPRMLQATGRYADAWIPIVLVRPTDYSCALEVVRTAASDAGRDPMSI
. : : ...:*. * : : :: . **: *
Mb0045c|M.bovis_AF2122/97 ERSAAV----DGGGLIASAEALAGLGVTLLTVGCDGPD------------
Rv0044c|M.tuberculosis_H37Rv ERSAAV----DGGGLIASAEALAGLGVTLLTVGCDGPD------------
MMAR_0063|M.marinum_M ERSAALQ---DSGRLSADADALAGLGVTLLTVGCDGPD------------
MUL_0062|M.ulcerans_Agy99 ERSAALQ---DSGRLSADADALAGLGVTLLTVGCDGPD------------
MAV_0061|M.avium_104 ERSAAVK---GGAELLANAEALVGLGVTLLTVGCDGPD------------
Mflv_0860|M.gilvum_PYR-GCK ERSAEAKGR-TEDALLENAAALTSLGVSMLTVGVNGPD------------
Mvan_6045|M.vanbaalenii_PYR-1 ERSAEAMGR-DADALIENADALHALGITMFTVGVNGPD------------
MSMEG_6907|M.smegmatis_MC2_155 ERSAGVQSDGGIEAMLAEADALADLGVTILTVGANGPD------------
TH_0517|M.thermoresistible__bu ERSSGLPVEGGVDAALAEAAALVEQGVTLLTVGVNGPD------------
MAB_3016c|M.abscessus_ATCC_199 ERSTSWGLDPERGLTAAEADAYVEVGARLFTVGIDADNG-----------
MLBr_00131|M.leprae_Br4923 TPAAVRG-IITGRTRDDVDEALDSVLVRMIALGVPGEAWARHGVEHPMGA
:: * ::::* .
Mb0045c|M.bovis_AF2122/97 -------------------------------------YDLSAAAALCRWR
Rv0044c|M.tuberculosis_H37Rv -------------------------------------YDLSAAAALCRWR
MMAR_0063|M.marinum_M -------------------------------------YDLAAAEALCRWR
MUL_0062|M.ulcerans_Agy99 -------------------------------------YDLAAAEALCRWR
MAV_0061|M.avium_104 -------------------------------------YDLRDAQALCKWR
Mflv_0860|M.gilvum_PYR-GCK -------------------------------------YDLSQAETLCRWR
Mvan_6045|M.vanbaalenii_PYR-1 -------------------------------------YDLAQAEVLCRWR
MSMEG_6907|M.smegmatis_MC2_155 -------------------------------------YDLTAAEALCRWR
TH_0517|M.thermoresistible__bu -------------------------------------YDLSIAEALCRWR
MAB_3016c|M.abscessus_ATCC_199 -------------------------------------YDLSVLDAPLAWR
MLBr_00131|M.leprae_Br4923 DFAGVQDIIPQTIDEETVVSYAAKVPAALMKEVLFSGTPEEVIDQVAEWR
**
Mb0045c|M.bovis_AF2122/97 DGR------------------------------------
Rv0044c|M.tuberculosis_H37Rv DGR------------------------------------
MMAR_0063|M.marinum_M DSR------------------------------------
MUL_0062|M.ulcerans_Agy99 DSR------------------------------------
MAV_0061|M.avium_104 DGR------------------------------------
Mflv_0860|M.gilvum_PYR-GCK DSHTAAG--------------------------------
Mvan_6045|M.vanbaalenii_PYR-1 DRRQAGD--------------------------------
MSMEG_6907|M.smegmatis_MC2_155 DGRHAN---------------------------------
TH_0517|M.thermoresistible__bu DNR------------------------------------
MAB_3016c|M.abscessus_ATCC_199 DSRS-----------------------------------
MLBr_00131|M.leprae_Br4923 DHGLKYLVVINGSLVNSSLRKTVSALLPHARVLRGLKKL
*