For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
PASSPPTARVVRIVEMLAAAMQPKTVSEIAASTGISRATATAVVNELEAAGWVARDDELRYRIGASLPRA VDTSMPREAQQLLARIAAEAQAGATISRISAKALTVVAKAQAGRRAVTGFAVGQSIPLAFPAGSAVMPWR AAEECEQWLGTAAGLSARSGQRLLQQVSDCGYVVYRPDHDDTGMVDLLSDLLGSVGSEVMEAALRERLLR QLSRLTSRPYSRDELTSDEVLPISYLAAPIFDGAEARYELQLGPLLPRATRVVRETLIATLMRGARELSV VLGDR*
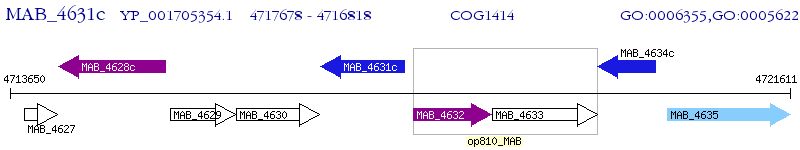
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_4631c | - | - | 100% (286) | IclR family transcriptional regulator |
| M. abscessus ATCC 19977 | MAB_4180 | - | 2e-12 | 29.24% (236) | IclR family transcriptional regulator |
| M. abscessus ATCC 19977 | MAB_3479c | - | 2e-10 | 25.97% (258) | IclR family regulatory protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_1528 | - | 2e-17 | 31.62% (253) | regulatory protein, IclR |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. marinum M | MMAR_3507 | - | 7e-13 | 28.05% (246) | IclR family transcriptional regulator |
| M. avium 104 | MAV_5001 | - | 6e-18 | 30.30% (264) | hypothetical protein MAV_5001 |
| M. smegmatis MC2 155 | MSMEG_5933 | - | 8e-14 | 26.98% (252) | hypothetical protein MSMEG_5933 |
| M. thermoresistible (build 8) | TH_4062 | - | 3e-11 | 27.65% (217) | PUTATIVE transcriptional regulator PadR family protein |
| M. ulcerans Agy99 | MUL_2762 | - | 1e-12 | 27.76% (245) | IclR family transcriptional regulator |
| M. vanbaalenii PYR-1 | Mvan_5230 | - | 7e-15 | 29.02% (255) | regulatory proteins, IclR |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1528|M.gilvum_PYR-GCK ------------------------MSVPVQPGRAS-----PPTDRVVQIL
Mvan_5230|M.vanbaalenii_PYR-1 ------------------------MSEPPAPGRAS-----PPTDRVVRVL
MSMEG_5933|M.smegmatis_MC2_155 ------------------------MLEQTDAGRAS-----PPTARVVAIL
MMAR_3507|M.marinum_M ------------------------MTDTVDPASVSSQGSSPPTERVIAVL
MUL_2762|M.ulcerans_Agy99 MPSPPALASVEISTYRSIDIPFDILTGTVDPASVSSQGLSPPTERVIAVL
MAV_5001|M.avium_104 ------------------------MAKSIGETPRRSGVKSPPTARVMDVL
TH_4062|M.thermoresistible__bu ------------------------MARPS-----------PQTERIVSLV
MAB_4631c|M.abscessus_ATCC_199 ------------------------MPASS-----------PPTARVVRIV
: * * *:: ::
Mflv_1528|M.gilvum_PYR-GCK DFLASRPTERYGVSDLARRVGLSKPTCLGIVTSLTEAGYLVR--DPADKT
Mvan_5230|M.vanbaalenii_PYR-1 DFLARHPQERFGVSELARRVDLTKPTCLGIVTSLRDAGYLVR--DPADKT
MSMEG_5933|M.smegmatis_MC2_155 DFLARHPHDRFGLSELSRRVGVSKPTCLGILTTLTESGYLVREGDGRDKC
MMAR_3507|M.marinum_M ELLGHNSAKQFSLAEICRGLGISRATGHAILTTLAAREWVIR--DPETFR
MUL_2762|M.ulcerans_Agy99 ELLGHNSAKQFSLAEICRGLGISRATGHAILTTLAAREWVIR--DPETFR
MAV_5001|M.avium_104 AAVADSPGG-LTSAELAKGCAISSSTCALVLAELERRAWVTR---RDDRR
TH_4062|M.thermoresistible__bu ELLRSQPRTGRSLADIARHLGVAKATCYPMVVALTEAGWLVR--HPTRKT
MAB_4631c|M.abscessus_ATCC_199 EMLAAAMQP-KTVSEIAASTGISRATATAVVNELEAAGWVAR---DDELR
: :::. :: .* :: * :: *
Mflv_1528|M.gilvum_PYR-GCK YRLGPSLITLGHRAQESMRVSPAARDHLRRLSARYGATAALSAVVDDRIT
Mvan_5230|M.vanbaalenii_PYR-1 YRLGPSLITLGHKAQESMRVSPAAREQLRRLSTRFGATAALSAVVDDRIT
MSMEG_5933|M.smegmatis_MC2_155 YRLGPALIALGHMAQESMRVNPFARAELRALSGAFDSTAAISAVIDDRIT
MMAR_3507|M.marinum_M YSWGPAIAGL-----DTPTTAQLHRGDLQELAAATGTQVMLARREDATLV
MUL_2762|M.ulcerans_Agy99 YSWGPAIAGL-----DTPTTAQLHRGDLQELAAATGTQVMLARREDATLV
MAV_5001|M.avium_104 YVLGSGLFGLVHGLREQFPLLDRGRDALRLLHERLGAGCSMSKIGARHLT
TH_4062|M.thermoresistible__bu YQLGPALVPIGAAAAAALDVLDLARPGMQELADTADMACVAFVPSGADLV
MAB_4631c|M.abscessus_ATCC_199 YRIGASLPRA-----VDTSMPREAQQLLARIAAEAQAGATISRISAKALT
* *..: : : : :.
Mflv_1528|M.gilvum_PYR-GCK LLDLVAPTGARP---GVEVGQSYPFAPPVGLMFVLWDD-EAEQDWLRRQP
Mvan_5230|M.vanbaalenii_PYR-1 LLDLVAPVGARP---GVEVGQSYPFAPPVGLMFVLWDD-EAVRDWLAMEP
MSMEG_5933|M.smegmatis_MC2_155 VLELVAPPGTDV---GVRVGQSYPFAPPVGLMYVLWDD-DALRAWLDKSP
MMAR_3507|M.marinum_M VIDTVGECPRGP---RISRGMRTPFVAPIGRDYVAWWSPDAQQDWLRAFG
MUL_2762|M.ulcerans_Agy99 VIDTVGECPRGP---RISRGMRTPFVAPIGRDYVAWWSPDAQQDWLRAFG
MAV_5001|M.avium_104 TVDSVG-HGTDG---EHAVGQRFPIDPPFGLVAMAWRDDDAVQAWLRRVT
TH_4062|M.thermoresistible__bu VSEIVQPAGGRRGTLGLRLGDRVQIIPPLGSVVAAWFSEDARWQWYRAGA
MAB_4631c|M.abscessus_ATCC_199 VVAKAQAGRRAVT--GFAVGQSIPLAFPAGSAVMPWRAAEECEQWLGTAA
. * : * * * : *
Mflv_1528|M.gilvum_PYR-GCK TIPLR---TDTERLKRVVASCRADGYLVERQTPGGRRLYALMAGMPTE-L
Mvan_5230|M.vanbaalenii_PYR-1 TVPLHPSRTDPDRLHRVIRRCRADGYLVERLTPGGRRLYSLMAGMSST-L
MSMEG_5933|M.smegmatis_MC2_155 TVPLR---TDSERLEKVIGECRNSGYLVERLTPAGQRLYGLMAGMSSR-L
MMAR_3507|M.marinum_M APNPR----FLQRMTLVLNQIQRRGFVVERLTRQYVRVYTALRALSADGE
MUL_2762|M.ulcerans_Agy99 APNPR----FLQRMILVLNQIQRRGFVVERLTRQYVRVYTALRALSADGE
MAV_5001|M.avium_104 PRLTR---TEIAQHQRVLADIRARGYGAWRFDDTHRSLHNRLAEALAS--
TH_4062|M.thermoresistible__bu EEFGVDPDAVEAAYGPVLELVRKRGFAVECLDQQERRLADAVAEVRGDGG
MAB_4631c|M.abscessus_ATCC_199 GLSAR-------SGQRLLQQVSDCGYVVYRPDHDDTGMVDLLSDLLGSVG
:: *: . : :
Mflv_1528|M.gilvum_PYR-GCK PDELRALLGELVSDIGERVYLR----EESSGRRKHDISVISAPVFDHYQR
Mvan_5230|M.vanbaalenii_PYR-1 PDELRALLGELVSDIGERVYLP----DEKPGRKRHDISVISAPVFDHYGR
MSMEG_5933|M.smegmatis_MC2_155 PDELQALLRELVSDIGERVHLR----SEATGRQRHDVSVISAPVHDHHQR
MMAR_3507|M.marinum_M VDEITTQLARAYADLAIIDVLD----EELTSGATHSVATISAPIRGPDGS
MUL_2762|M.ulcerans_Agy99 VDEITTQLARAYADLAIIDVLD----EELTSGATHSVATISAPIRGPDGS
MAV_5001|M.avium_104 -LEPTAQVARRLTTLMTMVTLRSV--TDVLETELPTTEFVVLPIFGHDGQ
TH_4062|M.thermoresistible__bu PLGRGRQTVTLRTVAGRPTVDVLI--GEIEPKRRYQPISINAAVFTAEAV
MAB_4631c|M.abscessus_ATCC_199 SEVMEAALRERLLRQLSRLTSRPYSRDELTSDEVLPISYLAAPIFDG-AE
: : .:
Mflv_1528|M.gilvum_PYR-GCK QVMVTSMHIG-KALTDNEISERARAVVAAADAVTAQLGGMRP--------
Mvan_5230|M.vanbaalenii_PYR-1 QVMVASMHIG-KALTDNEIRERAKALIGTADAVTAQLGGARPTSWPHTVG
MSMEG_5933|M.smegmatis_MC2_155 QVMVVSLHVG-RALTDSEITKRARGVVAAADAVTAQLGGVKPTR------
MMAR_3507|M.marinum_M ATMTVMATVF-ANLDGAGIRKLGERLRRSAHAIEQRSARSSA--------
MUL_2762|M.ulcerans_Agy99 ATMTVMATVF-ANLDGAGIRKLGERLRRSAHAIEQRSARSSA--------
MAV_5001|M.avium_104 PEYQIEIHLG-HSAG-LTLPELDDALEQARRLLTAPVR------------
TH_4062|M.thermoresistible__bu PALVLCLVDAPEPLTGRRVDDLGRRVASVAGRLTETLHGRVPDDRRVLRP
MAB_4631c|M.abscessus_ATCC_199 ARYELQLGPLLPRATRVVRETLIATLMRGARELSVVLGDR----------
: :
Mflv_1528|M.gilvum_PYR-GCK RDPD---
Mvan_5230|M.vanbaalenii_PYR-1 KGPDRDH
MSMEG_5933|M.smegmatis_MC2_155 -------
MMAR_3507|M.marinum_M -------
MUL_2762|M.ulcerans_Agy99 -------
MAV_5001|M.avium_104 -------
TH_4062|M.thermoresistible__bu ESR----
MAB_4631c|M.abscessus_ATCC_199 -------