For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VPADEDHQDAPPIDWRALLRQLPLLNILGVVAVLVAATLVVVKNLPDHGADQLLNVSYDPTRELYNVVDG TFTKQYKDKTGKTIEIKRTNGGSARQARSVLDGSQRADVVSLASVSDVDTLSLRGLIAPNWRQRLPNNSV PYTSTIVFVVRKGNPRGIHDWPDLIKENVSVITPNPRTSGNGQLSVLAAWGSVVTRGGTDTDARSFLTAL FRNVAALDSGARGATNTFSVQRLGDVHLTWENEAINEVDANKGELEIVYPPVSIRAEPAVAWVDANLGDP KRAAIARAYLEYLFTDEAQEVAAQHGYRPFKPDILARHSNTLPAIAQFPITAIASSWDDARQKFFSDNGI YETIPRNTDRGTTTFASDRQGR
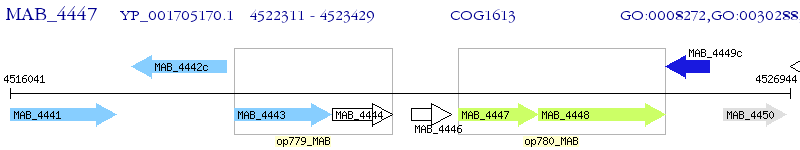
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_4447 | - | - | 100% (372) | Sulfate ABC transporter, periplasmic protein |
| M. abscessus ATCC 19977 | MAB_1652 | - | 7e-30 | 30.26% (271) | sulfate ABC transporter, sulfate-binding protein SubI |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2422c | subI | 3e-30 | 29.36% (327) | sulfate-binding lipoprotein |
| M. gilvum PYR-GCK | Mflv_2678 | - | 6e-29 | 32.35% (272) | sulfate ABC transporter, periplasmic sulfate-binding protein |
| M. tuberculosis H37Rv | Rv2400c | subI | 3e-30 | 29.36% (327) | sulfate-binding lipoprotein |
| M. leprae Br4923 | MLBr_00615 | subI | 6e-30 | 28.87% (336) | putative sulphate-binding protein |
| M. marinum M | MMAR_3718 | subI | 1e-31 | 34.08% (267) | sulfate-binding lipoprotein SubI |
| M. avium 104 | MAV_1782 | - | 7e-30 | 34.81% (270) | sulfate ABC transporter, sulfate-binding protein |
| M. smegmatis MC2 155 | MSMEG_4533 | - | 1e-31 | 32.72% (272) | sulfate-binding protein |
| M. thermoresistible (build 8) | TH_1447 | subI | 9e-31 | 29.52% (332) | PROBABLE SULFATE-BINDING LIPOPROTEIN SUBI |
| M. ulcerans Agy99 | MUL_3661 | subI | 2e-30 | 32.96% (267) | sulfate-binding lipoprotein SubI |
| M. vanbaalenii PYR-1 | Mvan_3875 | - | 2e-27 | 31.25% (272) | sulfate ABC transporter, periplasmic sulfate-binding protein |
CLUSTAL 2.0.9 multiple sequence alignment
MLBr_00615|M.leprae_Br4923 -------MFKDIGRFMRWRHSMVVGVTLASTTDVVAACHGGASDVVGG-G
MAV_1782|M.avium_104 ---------MDIRTAARWRP----VLALVLTAGVVAGCHGGASDAVGGTG
Mb2422c|M.bovis_AF2122/97 MLSLTLSEASCIASASRWRHIIPAGVVCALIAGIGVGCHGGPSDVVGRAG
Rv2400c|M.tuberculosis_H37Rv MLSLTLSEASCIASASRWRHIIPAGVVCALIAGIGVGCHGGPSDVVGRAG
MMAR_3718|M.marinum_M -------MPQTFMGAARSRRPGTFGVIFALIAVLASACQGGPSDIAGGGG
MUL_3661|M.ulcerans_Agy99 -------MPQTFMGAARSRRPGTFGVIFALIAVLASACQGGPSDIAGGGG
Mflv_2678|M.gilvum_PYR-GCK -------MLNPVRNLLKSPPRWRTAAALAVTTTLLAACGGGASDVAGDSG
Mvan_3875|M.vanbaalenii_PYR-1 -----------MRKILKSTRRWRTAAALTLSAAVLAACGGGASDVVGEEG
MSMEG_4533|M.smegmatis_MC2_155 ---------MPTTEKLTKFKFWRTATALAATGILAVGCGGGASDVAGDDG
TH_1447|M.thermoresistible__bu ----MSGQKSPRPARSLRRRGARTLSVLAAGALLLAGCGGGASDVPG--G
MAB_4447|M.abscessus_ATCC_1997 ---MPADEDHQDAPPIDWRALLRQLPLLNILGVVAVLVAATLVVVKNLPD
: . . .
MLBr_00615|M.leprae_Br4923 TAHA--HTSIALVAYSAPGPGWGKVIPAFNDA---RSGDGVQVVTSYGAS
MAV_1782|M.avium_104 PADA--HTSITLVAYSVPEPGWSKIIPAFNAS---DEGKGIQVVTSYGAS
Mb2422c|M.bovis_AF2122/97 PDRA--HTSITLVAYAVPEPGWSAVIPAFNAS---EQGRGVQVITSYGAS
Rv2400c|M.tuberculosis_H37Rv PDRA--HTSITLVAYAVPEPGWSAVIPAFNAS---EQGRGVQVITSYGAS
MMAR_3718|M.marinum_M PTDA--HTSITLVAYSAPEPGWSTVIPAFNAS---EEGKDVQVITSYGAS
MUL_3661|M.ulcerans_Agy99 PTDA--HTSITLVAYSAPEPGWSTVIPAFNAS---EEGKDVQVITSYGAS
Mflv_2678|M.gilvum_PYR-GCK GSGS-AETTLTLVAFAVPEPGWSKAAPAFSAT---EEGEGVEVTASYGAS
Mvan_3875|M.vanbaalenii_PYR-1 GNSD-AETTLTLVAFAVPEPGWSKVSPAFAAT---EEGKGVAVTGSYGAS
MSMEG_4533|M.smegmatis_MC2_155 GADANAETTLTLVAYAVPEPGWSKIIPAFTAT---DEGKGVAVRTSFGAS
TH_1447|M.thermoresistible__bu GGDADADTTLTLVAYAVPEPGWSKVIPAFAAT---EEGTGVQVRASYGAS
MAB_4447|M.abscessus_ATCC_1997 HGAD----QLLNVSYDPTRELYNVVDGTFTKQYKDKTGKTIEIKRTNGGS
: *:: . :. :* * : : : *.*
MLBr_00615|M.leprae_Br4923 GDQSRGVVDG-KPADIVNFSVETDITRLVKVGKVSKDWDTDA-TKGIPFG
MAV_1782|M.avium_104 GDQSRGVVDG-KPADVVNFSVEPDIARLVKAGKVAKDWNTDA-TKGIPFG
Mb2422c|M.bovis_AF2122/97 ADQSRGVADG-KPADLVNFSVEPDIARLVKAGKVDKDWDADA-TKGIPFG
Rv2400c|M.tuberculosis_H37Rv ADQSRGVADG-KPADLVNFSVEPDIARLVKAGKVDKDWDADA-TKGIPFG
MMAR_3718|M.marinum_M ADQSRGVADG-KPADLVNFSVEPDIARLVRAGKVAKDWDADA-TRGIPFE
MUL_3661|M.ulcerans_Agy99 ADQSRGVADG-KPADLVNFSVEPDIARLVRAGKVAKDWDAAA-TRGIPFE
Mflv_2678|M.gilvum_PYR-GCK GDQSRSVESG-KPADVVNFSVEPDITRLVKAGKVDENWNAGP-NKGIAFG
Mvan_3875|M.vanbaalenii_PYR-1 GDQSRAVESG-KPADIVNFSVEPDITRLVKAGKVDENWNAGP-NKGIAFG
MSMEG_4533|M.smegmatis_MC2_155 GDQSRAVVDG-KPADIVNFSVEPDVTRLVKAGKVAEDWNAGA-TKGIPFG
TH_1447|M.thermoresistible__bu GDQSRGVESG-KPADLVNFSVEPDITRLVEAGKVAEGWNANA-TKGIPFG
MAB_4447|M.abscessus_ATCC_1997 ARQARSVLDGSQRADVVSLASVSDVDTLSLRGLIAPNWRQRLPNNSVPYT
. *:*.* .* : **:*.:: .*: * * : .* ...:.:
MLBr_00615|M.leprae_Br4923 SVVTLVVRTGNPKNIKDWDDLVRPGVEVIMPNPLSSGSAKWNLLAPYAAK
MAV_1782|M.avium_104 SVVTLVVRKGNPKHIKDWDDLLRPGVEVITPSPLSSGSAKWNLLAPYAVK
Mb2422c|M.bovis_AF2122/97 SVVTFVVRAGNPKNIRDWDDLLRPGIEVITPSPLSSGSAKWNLLAPYAAK
Rv2400c|M.tuberculosis_H37Rv SVVTFVVRAGNPKNIRDWDDLLRPGIEVITPSPLSSGSAKWNLLAPYAAK
MMAR_3718|M.marinum_M SVVTLVVREGNPKNIRDWDDLLRPGVEVITPSPLSSGSAKWNLLAPYAVK
MUL_3661|M.ulcerans_Agy99 SVVTLVVREGNPKNIRNWDDLLRPGVEVITPSPLSSGSAKWNLLAPYAVK
Mflv_2678|M.gilvum_PYR-GCK SIVTFAVRPGNPKNIRTWDDLLQPGIEVITPSPLSSGAAKWNLLAPYAYA
Mvan_3875|M.vanbaalenii_PYR-1 SVVSFAVRPGNPKNIRTWDDLLQPGIEVITPSPLSSGAAKWNLLAPYAYA
MSMEG_4533|M.smegmatis_MC2_155 SVVSLVVRQGNPKGIKDWDDLLQPGLEVVTPSPLSSGSAKWNLLAPYAAK
TH_1447|M.thermoresistible__bu SVVTFMVREGNPKNIRDWDDLLRPDVEVITPSPLSSGSAKWNLLAPYAVK
MAB_4447|M.abscessus_ATCC_1997 STIVFVVRKGNPRGIHDWPDLIKENVSVITPNPRTSGNGQLSVLAAWGSV
* : : ** ***: *: * **:: .:.*: *.* :** .: .:**.:.
MLBr_00615|M.leprae_Br4923 SEGCRNSQAGIDFVNRLVAEHVKLRLGSAREATDAFVQGSGDVLISYENE
MAV_1782|M.avium_104 SEGGAHGDAGVDFIRKLVTEHVKLRPGSGREATDVFVQGSGDVLISYENE
Mb2422c|M.bovis_AF2122/97 SDGGRNNQAGIDFVNTLVNEHVKLRPGSGREATDVFVQGSGDVLISYENE
Rv2400c|M.tuberculosis_H37Rv SDGGRNNQAGIDFVNTLVNEHVKLRPGSGREATDVFVQGSGDVLISYENE
MMAR_3718|M.marinum_M SEGGRNSQAGIDFVSRLVREHVKLRPGSGREATDVFVQGSGDVLISYENE
MUL_3661|M.ulcerans_Agy99 SEGGRNSQAGIDFVSRLVREHVKLRPGSGREAIDVFVQGSGDVLISYENE
Mflv_2678|M.gilvum_PYR-GCK SNGGQNPEAGIEFVNKLVTEHVKLRPGSGREATDVFRQGSGDVLLAYENE
Mvan_3875|M.vanbaalenii_PYR-1 SNGGKDPQAGIDFVNKLVTEHVKLRPGSGREATDVFRQGSGDVLLAYENE
MSMEG_4533|M.smegmatis_MC2_155 SNGGQDPEAGIDFVNKLVTEHVKTRPGSGREATDVFLQGTGDVLISYENE
TH_1447|M.thermoresistible__bu SAGGQDRQAGLDYVTRLVNEHIKLRPGSGREATDVFLQGSGDVLLAYENE
MAB_4447|M.abscessus_ATCC_1997 VTRGGTDTDARSFLTALFRNVAALDSGARGATNTFSVQRLGDVHLTWENE
. .:: *. : *: : * *** :::***
MLBr_00615|M.leprae_Br4923 AIAAERQGK-PVEHINPPVTFKIENPLAVVST----SAHLEVATAFKNFQ
MAV_1782|M.avium_104 AIATERAGK-PVEHLNLAQTFKIDNPVAVVNT----SPHLQAAVAFKNFQ
Mb2422c|M.bovis_AF2122/97 AIATERAGK-PVQHVTPPQTFKIENPLAVVAT----STHLGAATAFRNFQ
Rv2400c|M.tuberculosis_H37Rv AIATERAGK-PVQHVTPPQTFKIENPLAVVAT----STHLGAATAFRNFQ
MMAR_3718|M.marinum_M AIATERQGK-RVQHVTPPQSFQIENPLAVVTT----TAHLATATAFKNFQ
MUL_3661|M.ulcerans_Agy99 AIATERQGK-RVQHVTPPQSFQIENPLAVVTT----TAHLATATAFKNFQ
Mflv_2678|M.gilvum_PYR-GCK ALNFD------LEHVNPAQTFKIENPTAVVNT----SRHLDKAQAFVDFQ
Mvan_3875|M.vanbaalenii_PYR-1 ALNFD------LEHVNPAQTFKIENPTAVVNT----SQHPDQAQAFVNFQ
MSMEG_4533|M.smegmatis_MC2_155 AIYVERQGK-PVEHVNPPQTFKIENPVAVVNT----TSHSEAANALKNFV
TH_1447|M.thermoresistible__bu AINARRQGH-RFDYLTPPQTFKIENPVAVVTT----SRHLDKATAFVNFQ
MAB_4447|M.abscessus_ATCC_1997 AINEVDANKGELEIVYPPVSIRAEPAVAWVDANLGDPKRAAIARAYLEYL
*: .: : . ::: : . * * : . : * * ::
MLBr_00615|M.leprae_Br4923 YTTAAQGLWAQAGFRPVDPAVSADFSDQFPAPVKLWTIADLGGWSTVDPQ
MAV_1782|M.avium_104 YTAAAQKVWAQAGFRPVDPAVAADFRDQYPVPAKLWTIADLGGWSAADPQ
Mb2422c|M.bovis_AF2122/97 YTVQAQKLWAQAGFRPVDPAVAADFADLFPVPAKLWTIADLGGWGSVDPQ
Rv2400c|M.tuberculosis_H37Rv YTVQAQKLWAQAGFRPVDPAVAADFADLFPVPAKLWTIADLGGWGSVDPQ
MMAR_3718|M.marinum_M YTVEAQRLWAQAGFRPVDPAVVAEFRDQFPLPEKLWTIADLGGWDTVDPQ
MUL_3661|M.ulcerans_Agy99 YTVEAQRLWAQAGFRPVDPAVVVEFRDQFPLPEKLWTIADLGGWDTVDPQ
Mflv_2678|M.gilvum_PYR-GCK FTPEGQKLWAEAGFRPVDPAVQAEFADKFPAPEKLWTIDDLGGWKNVDTQ
Mvan_3875|M.vanbaalenii_PYR-1 FTPEAQKLWAEANFRPVDPAVLAEFADKFPTPEKLWTIEDLGGWSKVDSE
MSMEG_4533|M.smegmatis_MC2_155 FTPEGQKIWAEAGFRPVDPAVAAEFAADFPAPEKLWTVDDLGGWDAVDPS
TH_1447|M.thermoresistible__bu YTPEAQRLWAEAGFRPVDPAVAADFADQFPAPEKLWTIDDLGGWEKVDVE
MAB_4447|M.abscessus_ATCC_1997 FTDEAQEVAAQHGYRPFKPDILARHSNTLPAIAQFPITAIASSWDDARQK
:* .* : *: .:**..* : . . * :: ..* . .
MLBr_00615|M.leprae_Br4923 LFDKN----------TGSITKVYMQATG-
MAV_1782|M.avium_104 LFDKN----------TGSITKIYTQATG-
Mb2422c|M.bovis_AF2122/97 LFDKA----------TGSITKIYLRATG-
Rv2400c|M.tuberculosis_H37Rv LFDKA----------TGSITKIYLRATG-
MMAR_3718|M.marinum_M LFDKG----------TGSITNIYMQATG-
MUL_3661|M.ulcerans_Agy99 LFDKG----------TGSITNIYMQATG-
Mflv_2678|M.gilvum_PYR-GCK LFDKD----------NGTITKIYKQATG-
Mvan_3875|M.vanbaalenii_PYR-1 LFDKE----------NGTITKIYKQATG-
MSMEG_4533|M.smegmatis_MC2_155 LFDKD----------NGTITKIYKQATG-
TH_1447|M.thermoresistible__bu LFNKD----------GGAITKIYIQATG-
MAB_4447|M.abscessus_ATCC_1997 FFSDNGIYETIPRNTDRGTTTFASDRQGR
:*.. *.. *