For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VRPRHGTCSQTNEGALINPQHAARLLGWMIPTGARLVFGVIFLLEGTQKVFGWWSGSPTGSGHPEPFLNW PYWWAGVFELVLGSLLTVGLRTRLAAVLAAGMMAYAYFFEHLQVHWEPMQNGGAFAATFCWGLLLLAVTA NDTRLSLDRVLARRA
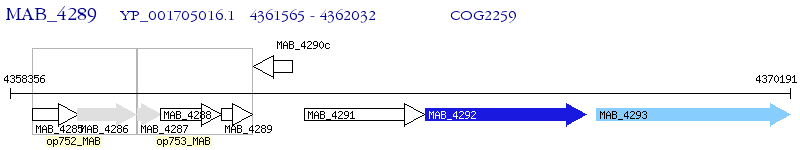
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_4289 | - | - | 100% (155) | hypothetical protein MAB_4289 |
| M. abscessus ATCC 19977 | MAB_1930c | - | 4e-20 | 40.16% (122) | hypothetical protein MAB_1930c |
| M. abscessus ATCC 19977 | MAB_1931c | - | 3e-19 | 43.70% (119) | hypothetical protein MAB_1931c |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3091c | - | 2e-18 | 43.64% (110) | integral membrane protein |
| M. gilvum PYR-GCK | Mflv_2914 | - | 9e-21 | 42.97% (128) | DoxX family protein |
| M. tuberculosis H37Rv | Rv3064c | - | 2e-18 | 43.64% (110) | integral membrane protein |
| M. leprae Br4923 | - | - | - | - | - |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | MAV_3854 | - | 8e-05 | 38.46% (78) | DoxX subfamily protein, putative |
| M. smegmatis MC2 155 | MSMEG_4292 | - | 4e-23 | 45.00% (120) | integral membrane protein |
| M. thermoresistible (build 8) | TH_0215 | - | 4e-23 | 40.46% (131) | integral membrane protein |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_3593 | - | 8e-28 | 50.00% (120) | DoxX family protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_2914|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_3593|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_4292|M.smegmatis_MC2_155 --------------------------------------------------
TH_0215|M.thermoresistible__bu --------------------------------------------------
Mb3091c|M.bovis_AF2122/97 --------------------------------------------------
Rv3064c|M.tuberculosis_H37Rv --------------------------------------------------
MAB_4289|M.abscessus_ATCC_1997 --------------------------------------------------
MAV_3854|M.avium_104 MTSQPNDAHWQRPGESPEPTPGRPASARLVDPEDDLTPVGYPGDFNPSTG
Mflv_2914|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_3593|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_4292|M.smegmatis_MC2_155 --------------------------------------------------
TH_0215|M.thermoresistible__bu -----------------------LIGPDRSDRIDRTGAAIRRAARPIWLY
Mb3091c|M.bovis_AF2122/97 --------------------------------------------------
Rv3064c|M.tuberculosis_H37Rv --------------------------------------------------
MAB_4289|M.abscessus_ATCC_1997 -------------------------------------MRPRHGTCSQTNE
MAV_3854|M.avium_104 TTTVIPYGGAAAVAGSGAAGYHLLEQQEPLPYVQPHSAARHAAPEPTDDD
Mflv_2914|M.gilvum_PYR-GCK ---MTT-FDARLASYSSPMLSIFRIMFGVLFTLHGTMKLFGWP-------
Mvan_3593|M.vanbaalenii_PYR-1 ---MTTNLDARLTSYSAPVLSIFRIVFGLLFTLHGAQKVLAWPTGMQAGP
MSMEG_4292|M.smegmatis_MC2_155 ---MTTNLDTRLATYSPVVVAIFRIVFGFLFAVHGASKLFAWPVDS----
TH_0215|M.thermoresistible__bu RLLMTTTVDARLARLSTPALALFRIIIGLLFALHGAMKLFGWP-------
Mb3091c|M.bovis_AF2122/97 ---MVKDLDRRLAGCLPAVLSLFRLVYGLLFAGYGSMILFGWPVT-----
Rv3064c|M.tuberculosis_H37Rv ---MVKDLDRRLAGCLPAVLSLFRLVYGLLFAGYGSMILFGWPVT-----
MAB_4289|M.abscessus_ATCC_1997 GALINPQHAARLLGWMIPTG--ARLVFGVIFLLEGTQKVFGWWSGSPTG-
MAV_3854|M.avium_104 DEHHDRLLDVGRRGTQHLGLLVLRAGLGVVLGAHGLQKLFGWWGGSGVTG
* *.:: * ::.*
Mflv_2914|M.gilvum_PYR-GCK -----LGAAVPVGTWPYWFAGLIEVVCGILITVGFFTRIAAFIAAGHMAV
Mvan_3593|M.vanbaalenii_PYR-1 GD--FTGPAVPVGTWPYWWAGLLEIVLGLLITVGLFTRIAAFIAAGQMAV
MSMEG_4292|M.smegmatis_MC2_155 -----GSGAVPVGTWPYWYAGVIELVLGLLIMVGLFTRIAAFIASGHMAV
TH_0215|M.thermoresistible__bu -----HGESVPVGTWPVWWAGLIELIVGLLVTVGLFTRISAIIGSGQMAV
Mb3091c|M.bovis_AF2122/97 -----SAQPVEFGSWPGWYAGVIELVAGLLIATGLFTRAVAFVASGEMAV
Rv3064c|M.tuberculosis_H37Rv -----SAQPVEFGSWPGWYAGVIELVAGLLIATGLFTRAVAFVASGEMAV
MAB_4289|M.abscessus_ATCC_1997 -----SGHPEPFLNWPYWWAGVFELVLGSLLTVGLRTRLAAVLAAGMMAY
MAV_3854|M.avium_104 LRNSLSDVGYQHADILAYVSAGGELVAGVLLVLGLFTPLAAAGALAFLIN
: :. *:: * *: *: * * . . :
Mflv_2914|M.gilvum_PYR-GCK AYFWQHWGIIGGEPKSFWPLGGDAGGNGGELAILYCFAFLLLATLG-AGA
Mvan_3593|M.vanbaalenii_PYR-1 AYFWQHL------PKAFFPIE-----NGGEPAILFCFGFLLLVAMG-AGA
MSMEG_4292|M.smegmatis_MC2_155 AYFWQHQ------PKGLLPLE-----NGGEPTVLFCFGFLLLFAIG-GGA
TH_0215|M.thermoresistible__bu AYFWMHWPPLEGEPTSFWPVV-----NGGEPALLYCFGFLLLAATG-PGA
Mb3091c|M.bovis_AF2122/97 AYFWMHQ------PYALWPIGGPPDGNGGTPAILFCFGFFLLVFTG-GGI
Rv3064c|M.tuberculosis_H37Rv AYFWMHQ------PYALWPIGGPPDGNGGTPAILFCFGFFLLVFTG-GGI
MAB_4289|M.abscessus_ATCC_1997 AYFFEHL------QVHWEPMQ-----NGGAFAATFCWGLLLLAVTANDTR
MAV_3854|M.avium_104 GLLATISARPHAHTFSFFLPQ------GHEYQITLIVMATAVILCG-PGR
. : * : .
Mflv_2914|M.gilvum_PYR-GCK WSVDGRRQVAGGSGVRRGRVVTGTAAPPVTAAPAAQPVRRGGLLSRFRRN
Mvan_3593|M.vanbaalenii_PYR-1 WSID---------ALRQGRAVVRS--------------------------
MSMEG_4292|M.smegmatis_MC2_155 FALD---------AARAKR-------------------------------
TH_0215|M.thermoresistible__bu WALD---------AMMRRRAPATA--------------------------
Mb3091c|M.bovis_AF2122/97 YSID------------ARRTVTA---------------------------
Rv3064c|M.tuberculosis_H37Rv YSID------------ARRTVTA---------------------------
MAB_4289|M.abscessus_ATCC_1997 LSLDR---------VLARRA------------------------------
MAV_3854|M.avium_104 YGLDAR-------RGWAHRPFIGSFVALLAGIAAGVGVWVALNGVNPIG-
.:* *
Mflv_2914|M.gilvum_PYR-GCK RY
Mvan_3593|M.vanbaalenii_PYR-1 --
MSMEG_4292|M.smegmatis_MC2_155 --
TH_0215|M.thermoresistible__bu --
Mb3091c|M.bovis_AF2122/97 --
Rv3064c|M.tuberculosis_H37Rv --
MAB_4289|M.abscessus_ATCC_1997 --
MAV_3854|M.avium_104 --