For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
APADIENKLTALTPITLSAFRVVTGFLFFLHGTSHLFGWPLASYLQPVGTLPWWAGIIEFVGGAAIALGI GTRLAAFVASGAMAVAYFTYHQPEGLLPIQNSGEAAALYSWSFLLLVFTGGGSLSLDRFITRAKR*
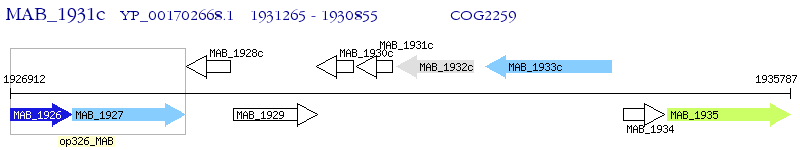
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_1931c | - | - | 100% (136) | hypothetical protein MAB_1931c |
| M. abscessus ATCC 19977 | MAB_1930c | - | 3e-41 | 64.75% (122) | hypothetical protein MAB_1930c |
| M. abscessus ATCC 19977 | MAB_4289 | - | 2e-19 | 43.70% (119) | hypothetical protein MAB_4289 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3091c | - | 4e-28 | 51.52% (132) | integral membrane protein |
| M. gilvum PYR-GCK | Mflv_2914 | - | 5e-25 | 46.97% (132) | DoxX family protein |
| M. tuberculosis H37Rv | Rv3064c | - | 4e-28 | 51.52% (132) | integral membrane protein |
| M. leprae Br4923 | - | - | - | - | - |
| M. marinum M | MMAR_4330 | - | 1e-05 | 27.87% (122) | hypothetical protein MMAR_4330 |
| M. avium 104 | MAV_1260 | - | 1e-05 | 27.56% (127) | DoxX subfamily protein, putative |
| M. smegmatis MC2 155 | MSMEG_4292 | - | 2e-33 | 51.11% (135) | integral membrane protein |
| M. thermoresistible (build 8) | TH_0215 | - | 3e-27 | 43.70% (135) | integral membrane protein |
| M. ulcerans Agy99 | MUL_0140 | - | 7e-06 | 27.87% (122) | hypothetical protein MUL_0140 |
| M. vanbaalenii PYR-1 | Mvan_3593 | - | 1e-27 | 44.78% (134) | DoxX family protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_2914|M.gilvum_PYR-GCK ------------------------------MTT-FDARLASYSSPMLSIF
Mvan_3593|M.vanbaalenii_PYR-1 ------------------------------MTTNLDARLTSYSAPVLSIF
MSMEG_4292|M.smegmatis_MC2_155 ------------------------------MTTNLDTRLATYSPVVVAIF
TH_0215|M.thermoresistible__bu LIGPDRSDRIDRTGAAIRRAARPIWLYRLLMTTTVDARLARLSTPALALF
Mb3091c|M.bovis_AF2122/97 ------------------------------MVKDLDRRLAGCLPAVLSLF
Rv3064c|M.tuberculosis_H37Rv ------------------------------MVKDLDRRLAGCLPAVLSLF
MAB_1931c|M.abscessus_ATCC_199 -----------------------------MAPADIENKLTALTPITLSAF
MMAR_4330|M.marinum_M ---------------------------------------MAPYDVGLLIL
MUL_0140|M.ulcerans_Agy99 ---------------------------------------MAPYDVGLLIL
MAV_1260|M.avium_104 ---------------------------------------MTPYDVGLLIL
: :
Mflv_2914|M.gilvum_PYR-GCK RIMFGVLFTLHGTMKLFGWP----------LGAAVPVGTWPYWFAGLIEV
Mvan_3593|M.vanbaalenii_PYR-1 RIVFGLLFTLHGAQKVLAWPTGMQAGPGDFTGPAVPVGTWPYWWAGLLEI
MSMEG_4292|M.smegmatis_MC2_155 RIVFGFLFAVHGASKLFAWPVDS-------GSGAVPVGTWPYWYAGVIEL
TH_0215|M.thermoresistible__bu RIIIGLLFALHGAMKLFGWP----------HGESVPVGTWPVWWAGLIEL
Mb3091c|M.bovis_AF2122/97 RLVYGLLFAGYGSMILFGWPVT--------SAQPVEFGSWPGWYAGVIEL
Rv3064c|M.tuberculosis_H37Rv RLVYGLLFAGYGSMILFGWPVT--------SAQPVEFGSWPGWYAGVIEL
MAB_1931c|M.abscessus_ATCC_199 RVVTGFLFFLHGTSHLFGWP---------LASYLQPVGTLP-WWAGIIEF
MMAR_4330|M.marinum_M RLVLGLTMAAHGFNKFFGGGRIPG-TARWFESIGMKPGKFHATVAAITEV
MUL_0140|M.ulcerans_Agy99 RLVLGLTMAAHGFNKFFGGGRIPG-TARWFESIGMKPGKFHATVAAITEV
MAV_1260|M.avium_104 RLVLGVTLAAHGYNKFFGGGRIPG-TARWFESIGMKPGKFHATVAASTEM
*:: *. : :* .:. . *. *. *.
Mflv_2914|M.gilvum_PYR-GCK VCGILITVGFFTRIAAFIAAGHMAVAYFWQHWGIIGGEPKSFWPLGGDAG
Mvan_3593|M.vanbaalenii_PYR-1 VLGLLITVGLFTRIAAFIAAGQMAVAYFWQHL------PKAFFPIE----
MSMEG_4292|M.smegmatis_MC2_155 VLGLLIMVGLFTRIAAFIASGHMAVAYFWQHQ------PKGLLPLE----
TH_0215|M.thermoresistible__bu IVGLLVTVGLFTRISAIIGSGQMAVAYFWMHWPPLEGEPTSFWPVV----
Mb3091c|M.bovis_AF2122/97 VAGLLIATGLFTRAVAFVASGEMAVAYFWMHQ------PYALWPIGGPPD
Rv3064c|M.tuberculosis_H37Rv VAGLLIATGLFTRAVAFVASGEMAVAYFWMHQ------PYALWPIGGPPD
MAB_1931c|M.abscessus_ATCC_199 VGGAAIALGIGTRLAAFVASGAMAVAYFTYHQ------PEGLLPIQ----
MMAR_4330|M.marinum_M SAGLGLAAGLLTPIPAAGFVSLMFVAAWTVHR------PNGFFIVK----
MUL_0140|M.ulcerans_Agy99 SAGLGLAAGLLTPIPAAGFVSLMFVAAWTVHR------PNGFFIVK----
MAV_1260|M.avium_104 AAGLGLAAGLLTPIPAAGFVSLMLVAAWTVHR------PNGFFIVK----
* : *: * * . * ** : * * .: :
Mflv_2914|M.gilvum_PYR-GCK GNGGELAILYCFAFLLLATLGAGAWSVDGRRQVAGGSGVRRGRVVTGTAA
Mvan_3593|M.vanbaalenii_PYR-1 -NGGEPAILFCFGFLLLVAMGAGAWSID---------ALRQGRAVVRS--
MSMEG_4292|M.smegmatis_MC2_155 -NGGEPTVLFCFGFLLLFAIGGGAFALD---------AARAKR-------
TH_0215|M.thermoresistible__bu -NGGEPALLYCFGFLLLAATGPGAWALD---------AMMRRRAPATA--
Mb3091c|M.bovis_AF2122/97 GNGGTPAILFCFGFFLLVFTGGGIYSID------------ARRTVTA---
Rv3064c|M.tuberculosis_H37Rv GNGGTPAILFCFGFFLLVFTGGGIYSID------------ARRTVTA---
MAB_1931c|M.abscessus_ATCC_199 -NSGEAAALYSWSFLLLVFTGGGSLSLD--------------RFITRAKR
MMAR_4330|M.marinum_M -EGWEYNLVLALSAVAVATIGAGKLSLDWVIFGNNWLNGWPGLVASLGLG
MUL_0140|M.ulcerans_Agy99 -EGWEYNLVLALSAVAVATIGAGKLSLDWVIFGNNWLNGWPGLVASLGLG
MAV_1260|M.avium_104 -EGWEYNLVLAASAVVVATLGAGRLSLDWLVFGKNWMDGWNGLLISLLLG
:. : . . . : * * ::*
Mflv_2914|M.gilvum_PYR-GCK PPVTAAPAAQPVRRGGLLSRFRRNRY
Mvan_3593|M.vanbaalenii_PYR-1 --------------------------
MSMEG_4292|M.smegmatis_MC2_155 --------------------------
TH_0215|M.thermoresistible__bu --------------------------
Mb3091c|M.bovis_AF2122/97 --------------------------
Rv3064c|M.tuberculosis_H37Rv --------------------------
MAB_1931c|M.abscessus_ATCC_199 --------------------------
MMAR_4330|M.marinum_M LAGAFGQLLIFYRPPARQAG------
MUL_0140|M.ulcerans_Agy99 LAGAFGQLLIFYRPPARRAG------
MAV_1260|M.avium_104 LAGAIGQLVIFYRPPAKQTG------