For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTVALVLMAGAALIGVAGPACLGPTVRPSLLPAAALATWLGALLAFLVLTTLSATLLLFPHVLDTSGVRT LVGGCLSGTVPHSSFGTLVQLGISLLPLTILARVAVVAIRSLRSARRRRARHFWMLRAASCRSGEVHWID DPRPIAYSLGGARGAVVATHGVRHLGERQCAAVIEHEMAHIRGRHHAAVLCADIAAAALPMLPLMRRAPA MVRLMVELAADDAAARVHGPPTVHAALLAMSQSAVAVPGVLHMSSESVAVRLLWLRTQGATGRTPMGRTR LMLGFALLPTTLAGSAVVALFRAYCTL
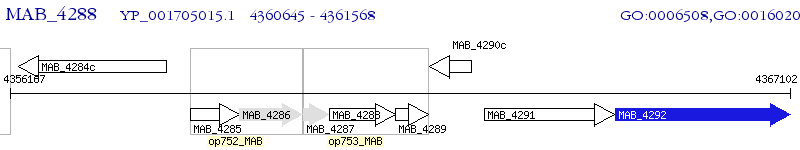
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_4288 | - | - | 100% (307) | hypothetical protein MAB_4288 |
| M. abscessus ATCC 19977 | MAB_4859 | - | 2e-18 | 26.28% (274) | hypothetical protein MAB_4859 |
| M. abscessus ATCC 19977 | MAB_2414c | - | 1e-17 | 27.93% (290) | hypothetical protein MAB_2414c |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1876c | - | 3e-22 | 31.83% (289) | hypothetical protein Mb1876c |
| M. gilvum PYR-GCK | Mflv_3358 | - | 2e-20 | 30.55% (275) | peptidase M48, Ste24p |
| M. tuberculosis H37Rv | Rv1845c | - | 3e-22 | 31.83% (289) | hypothetical protein Rv1845c |
| M. leprae Br4923 | MLBr_02064 | - | 1e-19 | 29.08% (282) | integral membrane protein |
| M. marinum M | MMAR_5059 | - | 4e-23 | 29.75% (316) | peptidase M48 like- protein |
| M. avium 104 | MAV_2870 | - | 4e-22 | 31.76% (296) | integral membrane protein |
| M. smegmatis MC2 155 | MSMEG_3631 | - | 1e-15 | 29.51% (244) | integral membrane protein |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_4135 | - | 2e-21 | 28.93% (318) | peptidase M48 like- protein |
| M. vanbaalenii PYR-1 | Mvan_5555 | - | 6e-23 | 32.75% (284) | peptidase M56, BlaR1 |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1876c|M.bovis_AF2122/97 -MSALAFTILAVLLAGPTPALLARATWPLRAPRAAMVLWQ-AIALAAVLS
Rv1845c|M.tuberculosis_H37Rv -MSALAFTILAVLLAGPTPALLARATWPLRAPRAAMVLWQ-AIALAAVLS
MLBr_02064|M.leprae_Br4923 -MSALAFTILAVLLVGPTPTLVARSTWPLRAPRAAMVLWQ-TIALAAALS
MAV_2870|M.avium_104 -MSALAFTILAVLLTGPVPAMLARARWPLRAPRAAMVLWQ-AVALAAVLS
Mflv_3358|M.gilvum_PYR-GCK -MSALAFSLLALLLSGPVPAMLARAAWPLRAPRAAIVLWQ-SIAAAAVLS
MSMEG_3631|M.smegmatis_MC2_155 -MSALAFTLVALALVGPVPAMLARASWPLRAPRAAIVLWQ-SIALAAVLS
MMAR_5059|M.marinum_M MNAALLLFAYASALIWVGPVLLRRVTGTGVHPRLAVASWL-SAVFAAIAA
MUL_4135|M.ulcerans_Agy99 MNAALLLFAYASALIWVGPVLLRRVTGTGVHPRLAVASWL-SAVFAAIAA
Mvan_5555|M.vanbaalenii_PYR-1 MNAVTVLLAYAVALSWFAPALLTASASAGAHPRLAVAGWI-AAVGSALSA
MAB_4288|M.abscessus_ATCC_1997 MTVALVLMAGAALIGVAGPACLGPTVRPSLLPAAALATWLGALLAFLVLT
. : * : *. : . * *:. * : :
Mb1876c|M.bovis_AF2122/97 SFSAGIAIASRLLMP-GPDGRPTTSFVGAAGRLGWPLWAAYITVFALTVL
Rv1845c|M.tuberculosis_H37Rv SFSAGIAIASRLLMP-GPDGRPTTSFVGAAGRLGWPLWAAYITVFALTVL
MLBr_02064|M.leprae_Br4923 TFSAGIAIASRVLMP-GPDGRLTASVIGAAGRLGWPLWAAYVAAFALTVL
MAV_2870|M.avium_104 AFSAGIAIATRVLVP-GPDGRPTTSILGAEGRLGWPLWTAYIGVFALTVL
Mflv_3358|M.gilvum_PYR-GCK AFSAGIAIASRLFVP-GPDGRPTSTIVGEIAVLGWPLWTAYVVVFLLTLV
MSMEG_3631|M.smegmatis_MC2_155 AFSAGIAIASRLFVP-GADGRPTATITSEIDVLGWPLWLLYVVVFSLTLF
MMAR_5059|M.marinum_M WVGALVFLGAAVLDS-IWRHR---ALALCLKELGLAGELGLPRGIASALA
MUL_4135|M.ulcerans_Agy99 WVGALVSLGAAVLDS-IWRHR---ALALCVKELGLAGELGLPRGIASALA
Mvan_5555|M.vanbaalenii_PYR-1 WAGAVVILVVGAVHA-VVTRS---ALTFCVETLGLTSAVHLPSSTATALV
MAB_4288|M.abscessus_ATCC_1997 TLSATLLLFPHVLDTSGVRTLVGGCLSGTVPHSSFGTLVQLGISLLPLTI
.* : : . . . .
Mb1876c|M.bovis_AF2122/97 VGARLAVAVVRVATATRRRRAHHRMVVDLVGVGHNGALAQPCARARDLRV
Rv1845c|M.tuberculosis_H37Rv VGARLAVAVVRVATATRRRRAHHRMVVDLVGVGHNGALAQPCARARDLRV
MLBr_02064|M.leprae_Br4923 VGARLIVAIVRVAIATRRRRAHHRMVVDLVGVGHNAALAQPCARARDLRV
MAV_2870|M.avium_104 VGARLMVAVVRVAIANRRRRAHHRMVVDLVGMGHGAALSQPCSRTRDLRV
Mflv_3358|M.gilvum_PYR-GCK IGARLVVSVLNVAIATRRRRAHHRMVVDLVG----AHHSRTRSRSS-LRV
MSMEG_3631|M.smegmatis_MC2_155 IGARLCVAIVQVGVATRRRRAHHRMMVDLLSKSRDAVPAHLRTPASGLRI
MMAR_5059|M.marinum_M VGVLSTGAVATVIACWRMGRQLRRQRSASRLHAATARTIGGASDWPGVVV
MUL_4135|M.ulcerans_Agy99 VGVLSTGAVATVIVCWRMGRQLRGQRSASRLHAATARTIGGVSDWPGVVV
Mvan_5555|M.vanbaalenii_PYR-1 VVLLIATSAVALYTVRRVVMALIRGRRRNVEHAEAVSIVGRPTEHPGVVE
MAB_4288|M.abscessus_ATCC_1997 LARVAVVAIRSLRSARRRRARHFWMLR------------AASCRSGEVHW
: : : * :
Mb1876c|M.bovis_AF2122/97 LDVAQPLAYCLPG-VRSRVVVSEGTLTALADAEVAAILTHERAHLRARHD
Rv1845c|M.tuberculosis_H37Rv LDVAQPLAYCLPG-VRSRVVVSEGTLTALADAEVAAILTHERAHLRARHD
MLBr_02064|M.leprae_Br4923 LEVAQPLAYCLPG-VRSRVVVSEGALTKLNDTEVTAILTHERAHLRARHD
MAV_2870|M.avium_104 LDVPQPLAYCLPG-VRSRVVVSEGTLSTLADAEVSAILTHERAHLRARHD
Mflv_3358|M.gilvum_PYR-GCK LDVDEPLAYCLPG-VRSRVVVSEGALKTLADNEMSAILEHEHAHLRARHD
MSMEG_3631|M.smegmatis_MC2_155 LDVAQPLAYCLPG-VRSRVVVSEGTLDTLSPDEVDAILTHERAHLRARHD
MMAR_5059|M.marinum_M VPAPQPAAYCVTGRPNSVIVVTSGALGRLDEPQLAAVLAHEYAHLASRHH
MUL_4135|M.ulcerans_Agy99 VPAPQPAAYCVTGRPNSVIVVTSGALGRLDEPQLAAVLAHEYAHLASRHH
Mvan_5555|M.vanbaalenii_PYR-1 ISARQPTAYCVSGGGRSVIVMTTAVRDLLDEDGMAAVLAHERAHLRGRHH
MAB_4288|M.abscessus_ATCC_1997 IDDPRPIAYSLGG-ARGAVVATHGVR-HLGERQCAAVIEHEMAHIRGRHH
: .* **.: * .. :* : .. * *:: ** **: .**.
Mb1876c|M.bovis_AF2122/97 LVLEAFTAVHAAFPRLVRSANALGAVQLLVELLADDAAVRAAGRTPLARA
Rv1845c|M.tuberculosis_H37Rv LVLEAFTAVHAAFPRLVRSANALGAVQLLVELLADDAAVRAAGRTPLARA
MLBr_02064|M.leprae_Br4923 LVLEAFTAVHAAFPRLVRSSAALSAVRLLVELLADDAAVRAAGRTPLARA
MAV_2870|M.avium_104 LVLEAFTAVHAAFPRLVRSANALGAVQLLVELLADDAAVRAAGRAPLARA
Mflv_3358|M.gilvum_PYR-GCK LVLEMFTAVHAAFPRLVRSGHALNAVRLLIELLADDAAVRAEGPTPLARA
MSMEG_3631|M.smegmatis_MC2_155 LVLEMFTAVHAAFPRFVRSASALDAVRLLIELLADDAAVRIAGPTPLARA
MMAR_5059|M.marinum_M DLLMALRALASSLPRLPLFAAAAKVVADLLEMCADDVAVRSHGKMALLRG
MUL_4135|M.ulcerans_Agy99 DLLMALRALASSLPRLPLFAAAAKVVADLLEMCADDVAVRSHGKMALLRG
Mvan_5555|M.vanbaalenii_PYR-1 HVIAVLNALAAALPRLPLMRRAAREVAALLEMCADDAASREHGRTHLLNS
MAB_4288|M.abscessus_ATCC_1997 AAVLCADIAAAALPMLPLMRRAPAMVRLMVELAADDAAARVHGPPTVHAA
: :::* : * * ::*: ***.* * * : .
Mb1876c|M.bovis_AF2122/97 LVACASG-RAPSGALAVGGPSTVLRVRRLSGRGNSAVLSAAAYLAAAAVL
Rv1845c|M.tuberculosis_H37Rv LVACASG-RAPSGALAVGGPSTVLRVRRLSGRGNSAVLSAAAYLAAAAVL
MLBr_02064|M.leprae_Br4923 LVACASG-QAPSGALAAGGNTTVLRVRRLSGRSNSAVVSAAAYLAAAIVL
MAV_2870|M.avium_104 LVACASG-RAPSGALAAGGPSTVVRVRRLGGRGNSPLLSAAAYLAAAAVF
Mflv_3358|M.gilvum_PYR-GCK LVACASG-RAPRGALAAGGPTTVVRVRRLGGKPNSRMLAAGAYAAAAAVL
MSMEG_3631|M.smegmatis_MC2_155 LVTCAAG-RTPSGALAAGGPTTVIRVRRLGGKPNSLTLSVIAYLAAVAIL
MMAR_5059|M.marinum_M LLALTEGPAMAPAALGAANTATVARAERLAISVSRRSWWQERLMLWATIG
MUL_4135|M.ulcerans_Agy99 LLALTEGPAMAPVALGAANTATVARAERLAISVSRRSWWQERLMLWATSG
Mvan_5555|M.vanbaalenii_PYR-1 LALLSTGDRLPEGALAAAGTAVLDRLLRLAQPPAT-SWWRPHVAVFAVMA
MAB_4288|M.abscessus_ATCC_1997 LLAMSQSAVAVPGVLHMSSESVAVRLLWLRTQGATGRTPMGRTRLMLGFA
* : . .* .. :. * *
Mb1876c|M.bovis_AF2122/97 VVPTVALAVPWLTQLQRLFIA--
Rv1845c|M.tuberculosis_H37Rv VVPTVALAVPWLTQLQRLFIA--
MLBr_02064|M.leprae_Br4923 LVPTVALAVPWLTELQRLFNI--
MAV_2870|M.avium_104 VVPTVALAVPWLTELERLLNL--
Mflv_3358|M.gilvum_PYR-GCK VVPTVAFAVPWLNELHRLFLV--
MSMEG_3631|M.smegmatis_MC2_155 VVPTVAVAVPWLTELHRLFSGLT
MMAR_5059|M.marinum_M LAVVSPMILGLACRL--------
MUL_4135|M.ulcerans_Agy99 LAVVSSMFLGLACRL--------
Mvan_5555|M.vanbaalenii_PYR-1 ACGVPIAALSLCVY---------
MAB_4288|M.abscessus_ATCC_1997 LLPTTLAGSAVVALFRAYCTL--
.