For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
ALNIADLIEHAIDTMPDRVAIISGDRKLTYAELEEQSNRLGHYLQSQGVGPGDKVGLYCRNGIEIVIALT AIVKIRAISVNVNYRYVEAELHYLFENSDMAALVHERRYSDKVANVLPSTPNVKTAIVVEDGADGSFDSY GGVPFADALAQGSPERDFEERSPDDIFLIYTGGTTGFPKGVMWRHEDIYRSLFGGINYVTGEYIEGEWDL AKQGAEAAPFIGFPIPPMIHGATQAATFMALFQGRTTVLAPEFNPEEVWELIEKHKINMLFFAGDAIGRP LIDALDTETGRARDLSSLWVLASSAALFSQTVKERYLELLPNRVITDAIGASETGTGGLSTVTKGQMHPG GPTVKISSTTTVLDEEGNPVQPGSGVRGLIAKSGHIPVGYFKDEKKTAETFKTFNGVRYAIPGDWATVEA DGTVTMLGRGSVSINTGGEKVFPEEVESVLKGHPAVFDAVVVGVPDEKWGQHVGAVIAVREGVELTFEDL DAHARKEIAGYKVPRSIWIVDSVKRNPAGKADYRWAKEVSETEKVDLVNSKHVNTGA*
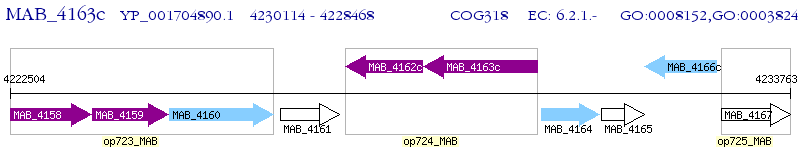
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_4163c | - | - | 100% (548) | acyl-CoA synthetase |
| M. abscessus ATCC 19977 | MAB_4559 | - | 2e-48 | 28.27% (520) | putative long-chain-fatty-acid CoA ligase |
| M. abscessus ATCC 19977 | MAB_4377c | - | 1e-42 | 29.23% (544) | AMP-binding domain protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3544c | fadD19 | 0.0 | 68.19% (547) | acyl-CoA synthetase |
| M. gilvum PYR-GCK | Mflv_1543 | - | 0.0 | 68.32% (546) | acyl-CoA synthetase |
| M. tuberculosis H37Rv | Rv3515c | fadD19 | 0.0 | 68.19% (547) | acyl-CoA synthetase |
| M. leprae Br4923 | MLBr_02546 | fadD2 | 5e-39 | 28.38% (511) | acyl-CoA synthetase |
| M. marinum M | MMAR_5001 | fadD19_1 | 0.0 | 67.40% (549) | fatty-acid-CoA ligase FadD19_1 |
| M. avium 104 | MAV_0644 | - | 0.0 | 69.72% (545) | acyl-CoA synthetase |
| M. smegmatis MC2 155 | MSMEG_5914 | - | 0.0 | 68.36% (550) | acyl-CoA synthetase |
| M. thermoresistible (build 8) | TH_1821 | fadD19 | 0.0 | 69.47% (547) | PROBABLE FATTY-ACID-CoA LIGASE FADD19 (FATTY-ACID-CoA |
| M. ulcerans Agy99 | MUL_4075 | fadD19_1 | 0.0 | 67.40% (549) | acyl-CoA synthetase |
| M. vanbaalenii PYR-1 | Mvan_5215 | - | 0.0 | 70.88% (546) | acyl-CoA synthetase |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_5914|M.smegmatis_MC2_155 ---------------------------------------MALN----IAD
Mvan_5215|M.vanbaalenii_PYR-1 ---------------------------------------MALN----IAD
Mflv_1543|M.gilvum_PYR-GCK ---------------------------------------MALN----IAD
Mb3544c|M.bovis_AF2122/97 -------------------------------------MAVALN----IAD
Rv3515c|M.tuberculosis_H37Rv -------------------------------------MAVALN----IAD
MMAR_5001|M.marinum_M -------------------------------------MAVALN----IAD
MUL_4075|M.ulcerans_Agy99 -------------------------------------MAVALN----IAD
MAV_0644|M.avium_104 -------------------------------------MAVALN----IAD
TH_1821|M.thermoresistible__bu ---------------------------------------VALN----IAD
MAB_4163c|M.abscessus_ATCC_199 ---------------------------------------MALN----IAD
MLBr_02546|M.leprae_Br4923 MTKLRHYLERGAAEVHYLRSIFESGALGFEPPLTYAAMAVAMRKWGDLAM
:*:. :*
MSMEG_5914|M.smegmatis_MC2_155 LAEHAIDAVPDRVALISGGDQLTYGQLEEKANRFAHYLIDQGVKKDDKVG
Mvan_5215|M.vanbaalenii_PYR-1 LAEHAIDAVPDRVALISGDEQLTYAELEEKSNRLAHYLIDQGVKKDDKVG
Mflv_1543|M.gilvum_PYR-GCK LAEHAIDAVPDRVAIITGDEQLTYAQLEEKANRLAHYLLDQGVKKDDKVG
Mb3544c|M.bovis_AF2122/97 LAEHAIDAVPDRVAVICGDEQLTYAQLEDKANRLAHHLIDQGVQKDDKVG
Rv3515c|M.tuberculosis_H37Rv LAEHAIDAVPDRVAVICGDEQLTYAQLEDKANRLAHHLIDQGVQKDDKVG
MMAR_5001|M.marinum_M LAEHAIDAVPDRVALICGDEQLTYAQLEEKANRLAHHLIDQGVGKGDKVG
MUL_4075|M.ulcerans_Agy99 LAEHAIDAVPDRVALICGDEQLTYAQLEEKANRLAHHLIDQGVRKGDKVG
MAV_0644|M.avium_104 LAEHAIDAVPDRVALICGDEKLTYAELEEKANRLAHYLLDQGVKKDDKVG
TH_1821|M.thermoresistible__bu LTEHAIDAVPDRVALICGDDQLTYAQLEEKANRLAHYLLDQGVRKDDKVG
MAB_4163c|M.abscessus_ATCC_199 LIEHAIDTMPDRVAIISGDRKLTYAELEEQSNRLGHYLQSQGVGPGDKVG
MLBr_02546|M.leprae_Br4923 LPAMPARRSPHRAALIDEEGTLTYKELDRAAHALANGLIAKGVRGGDGIA
* . *.*.*:* *** :*: :: :.: * :** .* :.
MSMEG_5914|M.smegmatis_MC2_155 LYCRNRIEIVIAMLGIVKAGAILVNVNFRYVEGELKYLFENSDMVALVHE
Mvan_5215|M.vanbaalenii_PYR-1 LYCRNRIEIVIAMLGIVKAGAILVNVNFRYVEGELKYLFENSDMVALVHE
Mflv_1543|M.gilvum_PYR-GCK LYCRNRNEIVIAMLGIVKAGAILVNVNFRYVEGELKYLFDNSDMVALVHE
Mb3544c|M.bovis_AF2122/97 LYCRNRIEIVIAMLGIVKAGAILVNVNFRYVEGELRYLFDNSDMVALVHE
Rv3515c|M.tuberculosis_H37Rv LYCRNRIEIVIAMLGIVKAGAILVNVNFRYVEGELRYLFDNSDMVALVHE
MMAR_5001|M.marinum_M LYCRNRIEIVIAMLGIIKAGAILINVNFRYVEGELKYLFDNSDMVALVHE
MUL_4075|M.ulcerans_Agy99 LYCRNRIEIVIAMLGIIKAGAILINVNFRYVEGELKYLFDNSDMVALVHE
MAV_0644|M.avium_104 LYCRNRNEIVIAMLGIVKAGAILVNVNYRYVEGELRYLFDNSDMVALVHE
TH_1821|M.thermoresistible__bu LYCRNRNEIVVAALACVKAGAIAVNVNFRYVEAELRYLFDNSDMVALVHE
MAB_4163c|M.abscessus_ATCC_199 LYCRNGIEIVIALTAIVKIRAISVNVNYRYVEAELHYLFENSDMAALVHE
MLBr_02546|M.leprae_Br4923 ILARNHRWFVIANYGCARVGARIILLNSEFSGPQIKEVSEREGAKVIIYD
: .** :*:* . : * : :* .: ::: : :... .::::
MSMEG_5914|M.smegmatis_MC2_155 RRYSDRVANVLPETPDIKTILVVEDGSDDDYERFGGVEFYSALEKSSPER
Mvan_5215|M.vanbaalenii_PYR-1 RRYADRVANVLPETPNVKTILVVEDGTDEDYQRYGGVEFYSALAQGSPER
Mflv_1543|M.gilvum_PYR-GCK RRYADRVANVLPETPKVKTILVIEDGSDDDYERYGGVEFNAALEQGSPER
Mb3544c|M.bovis_AF2122/97 RRYADRVANVLPDTPHVRTILVVEDGSDQDYRRYGGVEFYSAIAAGSPER
Rv3515c|M.tuberculosis_H37Rv RRYADRVANVLPDTPHVRTILVVEDGSDQDYRRYGGVEFYSAIAAGSPER
MMAR_5001|M.marinum_M RQYADRVANVLPDTPNVKTILVVQDGSDKDYRRYGGVEFYSAIADSSPER
MUL_4075|M.ulcerans_Agy99 RQYADRVANVLPDTPNVKTILVVQDGSDKDYQRYGGVEFYSAIADGSPER
MAV_0644|M.avium_104 RQYSDRVANVLPDTPNVKTILVVEDGSDKDYQRYGGVEFYSALEKGSPER
TH_1821|M.thermoresistible__bu RQYSDRVANVLPETPKVKTIVVIEDGSDLDYQRYGGVEFEAALAQGSPER
MAB_4163c|M.abscessus_ATCC_199 RRYSDKVANVLPSTPNVKTAIVVEDGADGSFDSYGGVPFADALAQGSPER
MLBr_02546|M.leprae_Br4923 DEYTKVVSKAEPALGKLRALGVNPDEEEPPGSTDETLAELIARSSSAPAP
.*:. *::. * .::: * * : : * .:*
MSMEG_5914|M.smegmatis_MC2_155 DFGP----RSEDDIYLLYTGGTTGFPKGVMWRHEDIYRVLFGGTDFATGE
Mvan_5215|M.vanbaalenii_PYR-1 DFGP----RSEDDIYLLYTGGTTGFPKGVMWRHEDIYRVLFGGTDFATGE
Mflv_1543|M.gilvum_PYR-GCK DFGP----RSEDDIYLLYTGGTTGFPKGVMWRHEDIYRVLFGGTDFATGE
Mb3544c|M.bovis_AF2122/97 DFGE----RSADAIYLLYTGGTTGFPKGVMWRHEDIYRVLFGGTDFATGE
Rv3515c|M.tuberculosis_H37Rv DFGE----RSADAIYLLYTGGTTGFPKGVMWRHEDIYRVLFGGTDFATGE
MMAR_5001|M.marinum_M DFAELQRERSADDIYILYTGGTTGFPKGVMWRHEDIYRVLFGGTDFATGE
MUL_4075|M.ulcerans_Agy99 DFAELQRERSADDIYILYTGGTTGFPKGVMWRHEDIYRVLFGGTDFATGE
MAV_0644|M.avium_104 DFGP----RSADDIYLLYTGGTTGFPKGVMWRHEDIYRVLFGGTDFATGE
TH_1821|M.thermoresistible__bu DFGP----RSADDIYLLYTGGTTGFPKGVMWRHEDIYRVLLGGIDFATGE
MAB_4163c|M.abscessus_ATCC_199 DFEE----RSPDDIFLIYTGGTTGFPKGVMWRHEDIYRSLFGGINYVTGE
MLBr_02546|M.leprae_Br4923 KVRK-------RASIIILTSGTTGTPKGANRNTPASLAPLGG--------
.. :: *.**** ***. . * *
MSMEG_5914|M.smegmatis_MC2_155 PIEDEYGLAKQAAANPPMVRYPIPPMIHGATQSATWMALFAGGTVLLTPE
Mvan_5215|M.vanbaalenii_PYR-1 PIADEYGLAKQAVESGPMVRYPIPPMIHGATQSATWMALFSGQTTVLTPE
Mflv_1543|M.gilvum_PYR-GCK PIADEYDLSRSAVDNPPMIRLPIPPMIHGATQSATWMALFSGHTVVLTPE
Mb3544c|M.bovis_AF2122/97 FVKDEYDLAKAAAANPPMIRYPIPPMIHGATQSATWMALFSGQTTVLAPE
Rv3515c|M.tuberculosis_H37Rv FVKDEYDLAKAAAANPPMIRYPIPPMIHGATQSATWMALFSGQTTVLAPE
MMAR_5001|M.marinum_M FIKDEYDLAKAAAANPPMIRYPIPPMIHGATQSATWMSIFSGQTTVLAPE
MUL_4075|M.ulcerans_Agy99 FIKDEYDLAKAAAANPPMIRYPIPPMIHGATQSATWMSIFSGQTTVLAPE
MAV_0644|M.avium_104 FVKDEYDLAKAAAENPPMIRYPIPPMIHGATQSATWMSIFSGQTTVLAPE
TH_1821|M.thermoresistible__bu YVADEYDHARQAKENPPMVRYPIPPMIHGATQSATWMALFGGGTVVLAPE
MAB_4163c|M.abscessus_ATCC_199 YIEGEWDLAKQGAEAAPFIGFPIPPMIHGATQAATFMALFQGRTTVLAPE
MLBr_02546|M.leprae_Br4923 -------ILSKVPFRAHEVTLLPAPMFHALGYLHATLAMFLGSTLVLRRR
: .**:*. : :::* * * :* .
MSMEG_5914|M.smegmatis_MC2_155 FNPDEVWQAIHDHKVNLLFFTGDAMARPLLDSLLAAKDAGKEYDLSSLFL
Mvan_5215|M.vanbaalenii_PYR-1 FDAEAVWRTIHEHKVNLLFFTGDAMARPLLDALLAAQDAGDQYDLSSLFL
Mflv_1543|M.gilvum_PYR-GCK FDADQIWRMIHEHKVNLLFFTGDAMARPLLDALLAHQEEGNTYDLSSLFL
Mb3544c|M.bovis_AF2122/97 FNADEVWRTIHKHKVNLLFFTGDAMARPLVDALVKGND----YDLSSLFL
Rv3515c|M.tuberculosis_H37Rv FNADEVWRTIHKHKVNLLFFTGDAMARPLVDALVKGND----YDLSSLFL
MMAR_5001|M.marinum_M FDADQVWRTISDRKVNLLFFTGDAMARPLLDALMKDND----YDLSSLFL
MUL_4075|M.ulcerans_Agy99 FDADQVWRTISDRKVNLLFFTGDAMARPLLDALMKDND----YDLSSLFL
MAV_0644|M.avium_104 FNADEVWRTIHEHKVNLVFFTGDAMARPLLDALNKDHD----YDLSSLFL
TH_1821|M.thermoresistible__bu FDPAEVWRTCEKHKVNLLFFTGDAMARPLVDALTAPGAN---YDLSNLFL
MAB_4163c|M.abscessus_ATCC_199 FNPEEVWELIEKHKINMLFFAGDAIGRPLIDALDTETGR--ARDLSSLWV
MLBr_02546|M.leprae_Br4923 FKPATVLEDIEKHQATAMVVVPVMLSR-ILDTLENLETK---SDLSSLRV
*.. : . .:: . :... :.* ::*:* ***.* :
MSMEG_5914|M.smegmatis_MC2_155 LASTAALFSTSLKEKFLELLPNRVITDSIGSSETGFGGTSIVAKGQSHTG
Mvan_5215|M.vanbaalenii_PYR-1 LASTAALFSTSLKEKFLELLPNRIITDSIGSSETGFGGTSIVAKGESHTG
Mflv_1543|M.gilvum_PYR-GCK LASTAALFSTSIKEKFLELLPNRIITDSIGSSETGFGGTSIVAKGESHTG
Mb3544c|M.bovis_AF2122/97 LASTAALFSPSIKEKLLELLPNRVITDSIGSSETGFGGTSVVAAGQAHGG
Rv3515c|M.tuberculosis_H37Rv LASTAALFSPSIKEKLLELLPNRVITDSIGSSETGFGGTSVVAAGQAHGG
MMAR_5001|M.marinum_M LASTAALFSPSIKEKLLELLPNRVITDSIGSSETGFGGTSIVGAGQATTG
MUL_4075|M.ulcerans_Agy99 LASTAALFSPSIKEKLLELLPNRVITDSIGSSETGFGGTSIVGAGQATTG
MAV_0644|M.avium_104 LASTAALFSPSIKERLLELLPNRVITDSIGSSETGFGGTSIVAKDAPHAG
TH_1821|M.thermoresistible__bu LASTAALFSPSLKDQLLELLPNRIITDSIGSSETGFGGSSVVTKGQTHTG
MAB_4163c|M.abscessus_ATCC_199 LASSAALFSQTVKERYLELLPNRVITDAIGASETGTGGLSTVTKGQMHPG
MLBr_02546|M.leprae_Br4923 VFVSGSQLGAELATRALEELG-PVIYNMYGSTEIAFATIAGPKDLQINSA
: :.: :. : : ** * :* : *::* . . : .
MSMEG_5914|M.smegmatis_MC2_155 GP-RVTIDKNTVVLDDDGNEVKPGSGVRGVIAKRGHIPLGYYKDEKKTAE
Mvan_5215|M.vanbaalenii_PYR-1 GP-RVTIDKNTVVLDEDGNPVTPGSGVRGIIAKRGHIPVGYFKDEKKTAE
Mflv_1543|M.gilvum_PYR-GCK GP-RVTIDKNTKVLDEDGNEVEPGSGKRGVIAKCGHIPVGYFKDEKKTAE
Mb3544c|M.bovis_AF2122/97 GP-RVRIDHRTVVLDDDGNEVKPGSGMRGVIAKKGNIPVGYYKDEKKTAE
Rv3515c|M.tuberculosis_H37Rv GP-RVRIDHRTVVLDDDGNEVKPGSGMRGVIAKKGNIPVGYYKDEKKTAE
MMAR_5001|M.marinum_M GP-RVTIDHRTVVLDEEGNEVKPGSGVRGIIAKKGNIPVGYYKDEKKTAE
MUL_4075|M.ulcerans_Agy99 GP-RVTIDHRTVVLDEEGNEVKPGSGVRGIIAKKGNIPVGYYKDEKKTAE
MAV_0644|M.avium_104 GP-RVTIDHRTVVLDEEGNEVKPGSGVRGLIAKKGNIPVGYYKDEKKTAE
TH_1821|M.thermoresistible__bu GP-RVTIDKNVVVLDEDGNEVKPGSGVRGLIAKRGHIPLGYYKDEKKTAE
MAB_4163c|M.abscessus_ATCC_199 GP-TVKISSTTTVLDEEGNPVQPGSGVRGLIAKSGHIPVGYFKDEKKTAE
MLBr_02546|M.leprae_Br4923 TVGPVVKGVKVKILDDSGKEVPRGQ-VGRIFVGNAFPFEGYTGGGGK---
* . . :**:.*: * *. ::. . ** . *
MSMEG_5914|M.smegmatis_MC2_155 TFKTINGVRYAIPGDYAEVEADGSVTMLGRGSVSINSGGEKIYPEEVEAA
Mvan_5215|M.vanbaalenii_PYR-1 TFKTINGVRYAIPGDYATVEADGSVTMLGRGSVSINSGGEKIYPEEVEAA
Mflv_1543|M.gilvum_PYR-GCK TFRTYNGVRYAIPGDYAEVEADGTVTMLGRGSVSINSGGEKIYPEEVEAA
Mb3544c|M.bovis_AF2122/97 TFRTINGVRYAIPGDYAQVEEDGTVTMLGRGSVSINSGGEKVYPEEVEAA
Rv3515c|M.tuberculosis_H37Rv TFRTINGVRYAIPGDYAQVEEDGTVTMLGRGSVSINSGGEKVYPEEVEAA
MMAR_5001|M.marinum_M TFKTINGVRYAIPGDYAMVEADGTVTMLGRGSVSINSGGEKIYPEEVEAA
MUL_4075|M.ulcerans_Agy99 TFKTINGVRYAIPGDYAMVEADGTVTMLGRGSVSINSGGEKIYPEEVEAA
MAV_0644|M.avium_104 TFKTFNGVRYAIPGDYALVEEDGTVTMLGRGSVSINSGGEKIYPEEVEGA
TH_1821|M.thermoresistible__bu TFKTINGVRYAIPGDWATVEADGTVTMLGRGSQSINSGGEKIYPEEVEGA
MAB_4163c|M.abscessus_ATCC_199 TFKTFNGVRYAIPGDWATVEADGTVTMLGRGSVSINTGGEKVFPEEVESV
MLBr_02546|M.leprae_Br4923 --QIIDGLLS--SGDVGYFDEHDLLYISGRDDEMIVSGGENVFPAEVEDL
: :*: .** . .: .. : : **.. * :***:::* ***
MSMEG_5914|M.smegmatis_MC2_155 LKGHPDVFDALVVGVPDPRFGQHVAAVVHPREGTRPTLAELDAHVRTEIA
Mvan_5215|M.vanbaalenii_PYR-1 LKGHPDVFDALVVGVPDPRFGQHVAAVVQPREGTRPALADLDAFVRTQIA
Mflv_1543|M.gilvum_PYR-GCK LKGHPDVFDALVVGVPDPRFGQHVAAVVHPRDGARPSLADLDAFVRKEIA
Mb3544c|M.bovis_AF2122/97 LKGHPDVFDALVVGVPDPRYGQQVAAVVQARPGCRPSLAELDSFVRSEIA
Rv3515c|M.tuberculosis_H37Rv LKGHPDVFDALVVGVPDPRYGQQVAAVVQARPGCRPSLAELDSFVRSEIA
MMAR_5001|M.marinum_M LKGHPDVFDALVVGVPDPRYGQHVAAVVAPRPGSRPSLAELDGFVRSAIA
MUL_4075|M.ulcerans_Agy99 LKGHPDVFDALVVGVPDPRYGQHVAAVVAPRPGSRPSLAELDGFVRSAIA
MAV_0644|M.avium_104 LKGHPDVFDALVVGVPDPRYGQHVAAVVQPRPGTRPSLAELDRFVRSEIA
TH_1821|M.thermoresistible__bu LKGHPDVFDALVVGVPDERFGQCVAAVVQRRPGTNPTLADLDAFVRQEIA
MAB_4163c|M.abscessus_ATCC_199 LKGHPAVFDAVVVGVPDEKWGQHVGAVIAVREGVELTFEDLDAHARKEIA
MLBr_02546|M.leprae_Br4923 ISGHPEVVEATAIGVDDKEWGARLRAFVVKKQDATIDEDAIKAYVRDHLA
:.*** *.:* .:** * .:* : *.: : . :. ..* :*
MSMEG_5914|M.smegmatis_MC2_155 GYKVPRSLWLVDEIKRSPAGKPDYRWAKDVTEERPADEVHANHVAANAK-
Mvan_5215|M.vanbaalenii_PYR-1 GYKVPRSLWLVDEVKRSPAGKPDYRWAKDVTEERPADDVHANHVGAK---
Mflv_1543|M.gilvum_PYR-GCK GYKVPRSLWLVDEIKRSPAGKPDYRWAKDTTEERPADEVHAKHVGAN---
Mb3544c|M.bovis_AF2122/97 GYKVPRSLWFVDEVKRSPAGKPDYRWAKEQTEARPADDVHAGHVTSGG--
Rv3515c|M.tuberculosis_H37Rv GYKVPRSLWFVDEVKRSPAGKPDYRWAKEQTEARPADDVHAGHVTSGG--
MMAR_5001|M.marinum_M GYKVPRSLWFVDEVKRSPAGKPDYRWAKEQTEARPADDVHAAHVSA----
MUL_4075|M.ulcerans_Agy99 GYKVPRSLWFVDEVKRSPAGKPDYRWAKEQTEARPADDVHAAHVSA----
MAV_0644|M.avium_104 GYKVPRSLWLVDEVKRSPAGKPDYRWAKEQTEARPADDVHAAHVSA----
TH_1821|M.thermoresistible__bu GYKVPRKIWWVDEIQRTPAGKPDYRWAKEQTEAREPDEVHSSHAGAGGGA
MAB_4163c|M.abscessus_ATCC_199 GYKVPRSIWIVDSVKRNPAGKADYRWAKEVSETEKVDLVNSKHVNTGA--
MLBr_02546|M.leprae_Br4923 RYKVPREVIFLEELPRNPTGKVLKRELRDFEVH-----------------
*****.: ::.: *.*:** * ::
MSMEG_5914|M.smegmatis_MC2_155 -
Mvan_5215|M.vanbaalenii_PYR-1 -
Mflv_1543|M.gilvum_PYR-GCK -
Mb3544c|M.bovis_AF2122/97 -
Rv3515c|M.tuberculosis_H37Rv -
MMAR_5001|M.marinum_M -
MUL_4075|M.ulcerans_Agy99 -
MAV_0644|M.avium_104 -
TH_1821|M.thermoresistible__bu K
MAB_4163c|M.abscessus_ATCC_199 -
MLBr_02546|M.leprae_Br4923 -