For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MPVDIKPVPKWLKYFNKVVIGGHKVGLKLPMMVLTVPGAKSGKPRSTPITPFTLDGQRYAVGGVPGSAWI TNARKAKTGTLTRGRHSEQVRIIELSPQDSRPVLRAFPIEVPAGVGFIRNAGLVTEGTPDEFEALAGIAA VFRFDPLNG
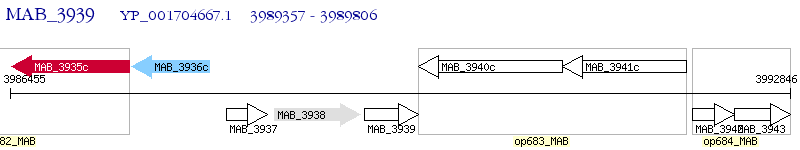
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_3939 | - | - | 100% (149) | hypothetical protein MAB_3939 |
| M. abscessus ATCC 19977 | MAB_3001 | - | 3e-42 | 57.55% (139) | hypothetical protein MAB_3001 |
| M. abscessus ATCC 19977 | MAB_2860c | - | 1e-06 | 35.80% (81) | hypothetical protein MAB_2860c |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_0024 | - | 3e-36 | 58.82% (119) | hypothetical protein Mflv_0024 |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | MLBr_02265 | - | 1e-07 | 45.83% (48) | hypothetical protein MLBr_02265 |
| M. marinum M | MMAR_2916 | - | 1e-37 | 54.68% (139) | hypothetical protein MMAR_2916 |
| M. avium 104 | MAV_4595 | - | 1e-44 | 60.99% (141) | cell entry (mce) related family protein |
| M. smegmatis MC2 155 | MSMEG_1077 | - | 2e-44 | 59.59% (146) | hypothetical protein MSMEG_1077 |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_2188 | - | 1e-37 | 54.68% (139) | hypothetical protein MUL_2188 |
| M. vanbaalenii PYR-1 | Mvan_0948 | - | 3e-42 | 60.56% (142) | hypothetical protein Mvan_0948 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_0024|M.gilvum_PYR-GCK --------------------MIAINRTGLLKN--GPVVLTVTGRRTGKPR
Mvan_0948|M.vanbaalenii_PYR-1 MASSERIRP-PWWLKYVNKVMIVLNRTGLFSD--GPVVLTVTGRKSGKPR
MSMEG_1077|M.smegmatis_MC2_155 -MRTERIRP-PWWLKYVNKVMIGLSKVGVGGDK-GPVVLTVPGRKTGKPR
MMAR_2916|M.marinum_M --MS-NPKP-PAWLKPMNKAMVTLQKIGVVAG--PVRVLTVTGRKTGLPR
MUL_2188|M.ulcerans_Agy99 --MS-NPKP-PAWLKPMNKAMVTLQKIGVVAG--PVRVLTVTGRKTGLPR
MAB_3939|M.abscessus_ATCC_1997 --MPVDIKPVPKWLKYFNKVVIGGHKVGLKL---PMMVLTVPGAKSGKPR
MAV_4595|M.avium_104 MGTSEHVRP-PWWLKPANKLFIRLSRLGLSFGGESPVVLTVPGRKSGAAR
MLBr_02265|M.leprae_Br4923 --------------------------------------------------
Mflv_0024|M.gilvum_PYR-GCK STPITPFEVDGRRYVVGGLPGSDWVRNLQANPDAVLTRGRTRERVRMVEL
Mvan_0948|M.vanbaalenii_PYR-1 STPITPFEVDGRRYVVGGLPGSDWVRNAQANPEAVLVRGRTREAVRMVEL
MSMEG_1077|M.smegmatis_MC2_155 TTPVTPMTVDGKRYVVGGLPGADWTANLRAAEEATIHQGRKAQRVRAVEM
MMAR_2916|M.marinum_M STPATPFVLDDCVYVVGGYPGADWVRNARAAGSGTVTRGRKKQQVVFTEL
MUL_2188|M.ulcerans_Agy99 STPATPFVLDDCVYVVGGYPGADWVRNARAAGSGTVTRGRKKQQVVFTEL
MAB_3939|M.abscessus_ATCC_1997 STPITPFTLDGQRYAVGGVPGSAWITNARKAKTGTLTRGRHSEQVRIIEL
MAV_4595|M.avium_104 STPVTPLTLDGRRYVVAGFPGADWVANARAAKQATIARGRHVERVRMTEL
MLBr_02265|M.leprae_Br4923 ---------------MPSLDQHTWT----------LPDG---------GA
: . * : *
Mflv_0024|M.gilvum_PYR-GCK PVEEARPLLRQFPVLVPTGVDFMKNAGLVTGPHPEEFEALAGRCPVFRFD
Mvan_0948|M.vanbaalenii_PYR-1 PTQQARPLLRRFPVLVPTGVGFMKNAGLVTGPHPDEFEALAGRCPVFRFD
MSMEG_1077|M.smegmatis_MC2_155 SVEEARPLLREFPILVPTGVGFMKNAGLVTGPHPDEFEALAGRCPVFRLD
MMAR_2916|M.marinum_M TAAQAVPVLAAFPHKVPRGVGFFKRSGLVRQGTAEEFAALAGQCAVFRLD
MUL_2188|M.ulcerans_Agy99 TAAQAVPVLAAFPHKVPRGVGFFKRSGLVRQGTAEEFAALAGQCAVFRLD
MAB_3939|M.abscessus_ATCC_1997 SPQDSRPVLRAFPIEVPAGVGFIRNAGLVTEGTPDEFEALAGIAAVFRFD
MAV_4595|M.avium_104 SAAEARPVLRAFPTEVPTGVGFMKRAGLVTDGRPEEFEALAGRCAVFRLD
MLBr_02265|M.leprae_Br4923 DAEDAKPLLRVWPSEVPTGVGCMKQFKLVNGGLTEKVEMLTGICPVVRFD
:: *:* :* ** **. ::. ** .::. *:* ..*.*:*
Mflv_0024|M.gilvum_PYR-GCK TV-----
Mvan_0948|M.vanbaalenii_PYR-1 TV-----
MSMEG_1077|M.smegmatis_MC2_155 PL-----
MMAR_2916|M.marinum_M PTAQN--
MUL_2188|M.ulcerans_Agy99 PTAQN--
MAB_3939|M.abscessus_ATCC_1997 PLNG---
MAV_4595|M.avium_104 PA-----
MLBr_02265|M.leprae_Br4923 SARFFFA
.