For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTTVTNTQPPRWLKPMNKVVRAFHRLGVSTGPAMVLTVPGRKTGQPRPTPITPFEVDGQLFAVGGYPGSD WARNAAAAGSGTLTKGNKVRHIRIVTVPPHVARRVLREFALQVPVGVRFAKSAGLVRQGTPDEFEGLAGT LSVFRFDPD
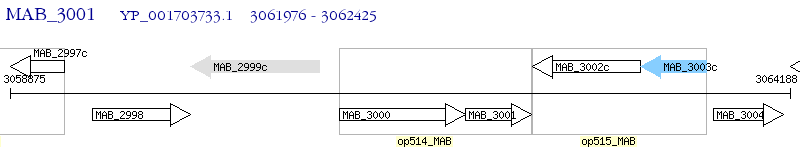
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_3001 | - | - | 100% (149) | hypothetical protein MAB_3001 |
| M. abscessus ATCC 19977 | MAB_3939 | - | 3e-42 | 57.55% (139) | hypothetical protein MAB_3939 |
| M. abscessus ATCC 19977 | MAB_3979c | - | 4e-07 | 29.66% (118) | hypothetical protein MAB_3979c |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0536c | - | 2e-06 | 32.20% (118) | hypothetical protein Mb0536c |
| M. gilvum PYR-GCK | Mflv_0024 | - | 7e-33 | 53.17% (126) | hypothetical protein Mflv_0024 |
| M. tuberculosis H37Rv | Rv0523c | - | 2e-06 | 32.20% (118) | hypothetical protein Rv0523c |
| M. leprae Br4923 | - | - | - | - | - |
| M. marinum M | MMAR_2916 | - | 1e-40 | 53.79% (145) | hypothetical protein MMAR_2916 |
| M. avium 104 | MAV_0161 | - | 5e-52 | 66.21% (145) | hypothetical protein MAV_0161 |
| M. smegmatis MC2 155 | MSMEG_1077 | - | 2e-36 | 52.45% (143) | hypothetical protein MSMEG_1077 |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_2188 | - | 1e-40 | 53.79% (145) | hypothetical protein MUL_2188 |
| M. vanbaalenii PYR-1 | Mvan_0948 | - | 1e-37 | 51.70% (147) | hypothetical protein Mvan_0948 |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_2916|M.marinum_M ---MSNPKPPAWLKPMN-KAMVTLQKIGVVAG-PVRVLTVTGRKTGLPRS
MUL_2188|M.ulcerans_Agy99 ---MSNPKPPAWLKPMN-KAMVTLQKIGVVAG-PVRVLTVTGRKTGLPRS
MAV_0161|M.avium_104 ---MTALRPPRYLKPMN-KVMMAVQRLGIPTG-PAMVLTVPGRKSGQPRS
Mflv_0024|M.gilvum_PYR-GCK --------------------MIAINRTGLLKN-GPVVLTVTGRRTGKPRS
Mvan_0948|M.vanbaalenii_PYR-1 MASSERIRPPWWLKYVN-KVMIVLNRTGLFSD-GPVVLTVTGRKSGKPRS
MSMEG_1077|M.smegmatis_MC2_155 -MRTERIRPPWWLKYVN-KVMIGLSKVGVGGDKGPVVLTVPGRKTGKPRT
MAB_3001|M.abscessus_ATCC_1997 MTTVTNTQPPRWLKPMN-KVVRAFHRLGVSTG-PAMVLTVPGRKTGQPRP
Mb0536c|M.bovis_AF2122/97 ------MQLPQWLARFNRYVTNPIQRLWAGWLPAFAILEHVGRRSGKPYR
Rv0523c|M.tuberculosis_H37Rv ------MQLPQWLARFNRYVTNPIQRLWAGWLPAFAILEHVGRRSGKPYR
. : :* **::* *
MMAR_2916|M.marinum_M TPATPFVLD-----DCVYVVGGYPGADWVRNARAAGSGTVTRGRKKQQVV
MUL_2188|M.ulcerans_Agy99 TPATPFVLD-----DCVYVVGGYPGADWVRNARAAGSGTVTRGRKKQQVV
MAV_0161|M.avium_104 TPMTPFEFR-----GGLYVVAGYPGADWAANARAAGTGTLRRGRRSRRVR
Mflv_0024|M.gilvum_PYR-GCK TPITPFEVD-----GRRYVVGGLPGSDWVRNLQANPDAVLTRGRTRERVR
Mvan_0948|M.vanbaalenii_PYR-1 TPITPFEVD-----GRRYVVGGLPGSDWVRNAQANPEAVLVRGRTREAVR
MSMEG_1077|M.smegmatis_MC2_155 TPVTPMTVD-----GKRYVVGGLPGADWTANLRAAEEATIHQGRKAQRVR
MAB_3001|M.abscessus_ATCC_1997 TPITPFEVD-----GQLFAVGGYPGSDWARNAAAAGSGTLTKGNKVRHIR
Mb0536c|M.bovis_AF2122/97 TPLNVFSADVDGRAGVAILLTYGPNRDWLKNITAAGGGRMRRYG------
Rv0523c|M.tuberculosis_H37Rv TPLNVFSADVDGRAGVAILLTYGPNRDWLKNITAAGGGRMRRYG------
** . : . : *. ** * * . : :
MMAR_2916|M.marinum_M FTELTAAQAVPVLAAFPHKVPRGVGFFKRSGLVRQGTAEEFAALAGQCAV
MUL_2188|M.ulcerans_Agy99 FTELTAAQAVPVLAAFPHKVPRGVGFFKRSGLVRQGTAEEFAALAGQCAV
MAV_0161|M.avium_104 IVELSADEARPVLREFPAKVPVGVGFAKRSGLVRDGTADEFEALAGQLAV
Mflv_0024|M.gilvum_PYR-GCK MVELPVEEARPLLRQFPVLVPTGVDFMKNAGLVTGPHPEEFEALAGRCPV
Mvan_0948|M.vanbaalenii_PYR-1 MVELPTQQARPLLRRFPVLVPTGVGFMKNAGLVTGPHPDEFEALAGRCPV
MSMEG_1077|M.smegmatis_MC2_155 AVEMSVEEARPLLREFPILVPTGVGFMKNAGLVTGPHPDEFEALAGRCPV
MAB_3001|M.abscessus_ATCC_1997 IVTVPPHVARRVLREFALQVPVGVRFAKSAGLVRQGTPDEFEGLAGTLSV
Mb0536c|M.bovis_AF2122/97 -------------KTFGVANPRRLTKAEAAPYVSSRWRPVFARLPFDEAV
Rv0523c|M.tuberculosis_H37Rv -------------KTFGVANPRRLTKAEAAPYVSSRWRPVFARLPFDEAV
* * : : : * * *. .*
MMAR_2916|M.marinum_M FRLDPTAQN
MUL_2188|M.ulcerans_Agy99 FRLDPTAQN
MAV_0161|M.avium_104 FRFDPA---
Mflv_0024|M.gilvum_PYR-GCK FRFDTV---
Mvan_0948|M.vanbaalenii_PYR-1 FRFDTV---
MSMEG_1077|M.smegmatis_MC2_155 FRLDPL---
MAB_3001|M.abscessus_ATCC_1997 FRFDPD---
Mb0536c|M.bovis_AF2122/97 LLTKAD---
Rv0523c|M.tuberculosis_H37Rv LLTKAD---
: ..