For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SKLIEFNETARRALEAGVNKLADAVKVTLGPRGRHVVLAKAFGGPAVTNDGVTIARDIDLEDPFENLGAQ LVKSVATKTNDVAGDGTTTATVLAQALVRAGLRNVAAGANPIALGAGISRAADAVSERLLTVAAPVAGEH AIAQVATVSSRDPEIGELVGEAMTKVGGDGVVSVEESSTINTELVITDGVQFDKGYLSQYFVTDFDDQKA VLEDALVLLHRDKISSLPDLLPLLELVAKEGKPLLIVAEDVEGEPLSTLVVNAIRKTLKAVAVKAPFFGD RRKAFLEDLAIVTNAQVVNPDVGLSLREVGIEVLGTARRVVVSKDETVIVDGGGSEDAIKARVAQLKAEI ETTDSDWDREKLQERLAKLAGGVAVIKVGAPTDTALKERKHRVEDAVSAAKAAVEEGIVAGGGSALVQAR SALDGLRAELKGDEAKGVDVFEAALSAPLFWIATNAGLDGAVVVSKVAELDAGHGFNAATLEYGDLIADG VIDPVKVTRSAVINAASAARMILTTETSIVDKPAEEADHGHGHHGHAH*
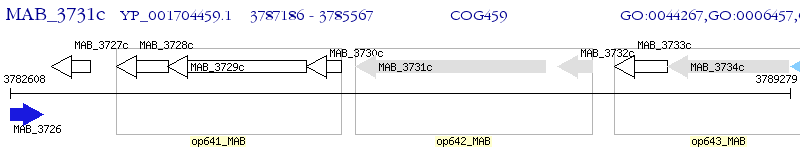
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_3731c | - | - | 100% (539) | 60 kDa chaperonin 1 (GroEL protein 1) |
| M. abscessus ATCC 19977 | MAB_0650 | groEL | e-180 | 63.07% (528) | chaperonin GroEL |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3451c | groEL | 0.0 | 77.96% (540) | chaperonin GroEL |
| M. gilvum PYR-GCK | Mflv_4924 | groEL | 0.0 | 82.59% (540) | chaperonin GroEL |
| M. tuberculosis H37Rv | Rv3417c | groEL | 0.0 | 77.96% (540) | chaperonin GroEL |
| M. leprae Br4923 | MLBr_00381 | groEL | 0.0 | 78.11% (539) | chaperonin GroEL |
| M. marinum M | MMAR_1126 | groEL | 0.0 | 79.22% (539) | 60 kDa chaperone (GroEL1) |
| M. avium 104 | MAV_4365 | groEL | 0.0 | 81.41% (538) | chaperonin GroEL |
| M. smegmatis MC2 155 | MSMEG_1583 | groEL | 0.0 | 83.15% (540) | chaperonin GroEL |
| M. thermoresistible (build 8) | TH_1687 | groEL1 | 0.0 | 81.31% (535) | 60 KDA CHAPERONIN 1 GROEL1 (PROTEIN CPN60-1) (GROEL |
| M. ulcerans Agy99 | MUL_0886 | groEL | 0.0 | 78.66% (539) | chaperonin GroEL |
| M. vanbaalenii PYR-1 | Mvan_1495 | groEL | 0.0 | 81.43% (544) | chaperonin GroEL |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3451c|M.bovis_AF2122/97 MSKLIEYDETARRAMEVGMDKLADTVRVTLGPRGRHVVLAKAFGGPTVTN
Rv3417c|M.tuberculosis_H37Rv MSKLIEYDETARRAMEVGMDKLADTVRVTLGPRGRHVVLAKAFGGPTVTN
MMAR_1126|M.marinum_M MSKLIEYDETARRAMEAGVDKLADTVRVTLGPRGRHVVLAKSFGGPTVTN
MUL_0886|M.ulcerans_Agy99 MSKLIEYDETARRAMEAGVDKLAETVRVTLGPRGRHVVLAKSFGGPTVTN
MLBr_00381|M.leprae_Br4923 MSKLIEYDETARHAMEVGMNKLADTVRVTLGPRGRHVVLAKAFGGPTITN
MAV_4365|M.avium_104 MSKIIEYDETARRAIEAGVNTLADAVRVTLGPRGRHVVLAKAFGGPAVTN
Mflv_4924|M.gilvum_PYR-GCK MSKQIEFNETARRAMEAGVDKLADAVKVTLGPRGRHVVLAKSWGGPTVTN
Mvan_1495|M.vanbaalenii_PYR-1 MSKQIEFNETARRAMEIGVDKLADAVKVTLGPRGRHVVLAKSWGGPTVTN
MSMEG_1583|M.smegmatis_MC2_155 MSKQIEFNETARRAMEAGVDKLADAVKVTLGPRGRHVVLAKSFGGPQVTN
TH_1687|M.thermoresistible__bu MSKLIEYNEIARRALETGVDKLADAVKVTLGPRGRHVVLAKAFGGPQVTN
MAB_3731c|M.abscessus_ATCC_199 MSKLIEFNETARRALEAGVNKLADAVKVTLGPRGRHVVLAKAFGGPAVTN
*** **::* **:*:* *::.**::*:**************::*** :**
Mb3451c|M.bovis_AF2122/97 DGVTVAREIELEDPFEDLGAQLVKSVATKTNDVAGDGTTTATILAQALIK
Rv3417c|M.tuberculosis_H37Rv DGVTVAREIELEDPFEDLGAQLVKSVATKTNDVAGDGTTTATILAQALIK
MMAR_1126|M.marinum_M DGVTVARDIDLEDPFENLGAQLVKSVATKTNDVAGDGTTTATVLAQALVK
MUL_0886|M.ulcerans_Agy99 DGVTVARDIDLEDPFENLGAQLVKSVATKTNDVAGDGTTTATVLARALVK
MLBr_00381|M.leprae_Br4923 DGVTVAREIDLEDPFENLGAQLVKSVATKTNDVAGDGTTTATVLAQALVK
MAV_4365|M.avium_104 DGVTVAREIDLEDPFENLGAQLVKSVATKTNDVAGDGTTTATVLAQALVK
Mflv_4924|M.gilvum_PYR-GCK DGVTIAREIDLEDPFENLGAQLVKSVATKTNDVAGDGTTTATVLAQALVK
Mvan_1495|M.vanbaalenii_PYR-1 DGVTIAREIDLEDPFENLGAQLVKSVATKTNDVAGDGTTTATVLAQAIIK
MSMEG_1583|M.smegmatis_MC2_155 DGVTIAREIDLEDPYENLGAQLVKSVATKTNDVAGDGTTTATVLAQALVR
TH_1687|M.thermoresistible__bu DGVTIAREIELEDPFENLGAQLVKSVATKTNDIAGDGTTTATVLAQALVQ
MAB_3731c|M.abscessus_ATCC_199 DGVTIARDIDLEDPFENLGAQLVKSVATKTNDVAGDGTTTATVLAQALVR
****:**:*:****:*:***************:*********:**:*:::
Mb3451c|M.bovis_AF2122/97 GGLRLVAAGVNPIALGVGIGKAADAVSEALLASATPVSGKTGIAQVATVS
Rv3417c|M.tuberculosis_H37Rv GGLRLVAAGVNPIALGVGIGKAADAVSEALLASATPVSGKTGIAQVATVS
MMAR_1126|M.marinum_M TGLRLVAAGINPIALGSGIGKAADAVSEALLASATPVSGKDAIAQVATVS
MUL_0886|M.ulcerans_Agy99 TGLRLVAAGINPIALGSGIGKAADAVSEALLASATPVSGKDAIAQVATVS
MLBr_00381|M.leprae_Br4923 GGLRMVAAGANPVALGAGISKAADAVSEALLAVATPVAGKDAITQVATVS
MAV_4365|M.avium_104 GGLRLVAAGANPIELGAGISKAADAVSEALLASATTVSGKDAIAQVATVS
Mflv_4924|M.gilvum_PYR-GCK AGLRNVAAGANPIALGAGIAKAADAVSEALLASATPVKDAKSIGQVATVS
Mvan_1495|M.vanbaalenii_PYR-1 GGLRNVAAGANPIALGAGISKAADAVSEALLAGATPVTDKKSIAQVATVS
MSMEG_1583|M.smegmatis_MC2_155 AGLRNVAAGANPIALGSGISKAADAVSEALLASATPVDDKKAIAQVATVS
TH_1687|M.thermoresistible__bu AGLKNVAAGANPLALGSGIAKAADAVGEALLAAATPVKDRSAIAQVATVS
MAB_3731c|M.abscessus_ATCC_199 AGLRNVAAGANPIALGAGISRAADAVSERLLTVAAPVAGEHAIAQVATVS
**: **** **: ** **.:*****.* **: *:.* . .* ******
Mb3451c|M.bovis_AF2122/97 SRDEQIGDLVGEAMSKVGHDGVVSVEESSTLGTELEFTEGIGFDKGFLSA
Rv3417c|M.tuberculosis_H37Rv SRDEQIGDLVGEAMSKVGHDGVVSVEESSTLGTELEFTEGIGFDKGFLSA
MMAR_1126|M.marinum_M SRDQLIGDLVGEAMSKVGHDGVVSVEESSTLGTELEFTEGVGFDKGYLSA
MUL_0886|M.ulcerans_Agy99 SRDQLIGDLVGEAMSKVGHDGVVSVEESSTLGTELEFTEGVGFDKGYLSA
MLBr_00381|M.leprae_Br4923 SRDEQIGALVGEGMNKVGTDGVVSVEESSTLDTELEFTEGVGFDKGFLSA
MAV_4365|M.avium_104 SRDQVLGELVGEAMTKVGVDGVVSVEESSTLNTELEFTEGVGFDKGFLSA
Mflv_4924|M.gilvum_PYR-GCK SRDELVGELVGEAMTKVGTDGVVTVEESSTLNTELEVTEGVGFDKGFISA
Mvan_1495|M.vanbaalenii_PYR-1 SRDEEVGELVGEAMTRVGHDGVVTVEESSTLLTELEITEGVGFDKGFLSA
MSMEG_1583|M.smegmatis_MC2_155 SRDEQVGELVGEAMTKVGHDGVVTVEESSTLETYLEVTEGVGFDKGFLSA
TH_1687|M.thermoresistible__bu SRDPEIGEMVAEAMTRVGHDGVVTVEESSTLDTEVEITEGVGFDKGYLSA
MAB_3731c|M.abscessus_ATCC_199 SRDPEIGELVGEAMTKVGGDGVVSVEESSTINTELVITDGVQFDKGYLSQ
*** :* :*.*.*.:** ****:******: * : .*:*: ****::*
Mb3451c|M.bovis_AF2122/97 YFVTDFDNQQAVLEDALILLHQDKISSLPDLLPLLEKVAGTGKPLLIVAE
Rv3417c|M.tuberculosis_H37Rv YFVTDFDNQQAVLEDALILLHQDKISSLPDLLPLLEKVAGTGKPLLIVAE
MMAR_1126|M.marinum_M YFVTDFDAQQAVLEDPLILLHQDKISSLPDLLPLLEKVAESGKPLMIIAE
MUL_0886|M.ulcerans_Agy99 YFVTDFDAQQAVLEDPLILLHQDKISSLPDLLPLLEKVAESGKPLMIIAE
MLBr_00381|M.leprae_Br4923 YFVTDFDSQQAVLDDPLVLLHQEKISSLPELLPMLEKVTESGKPLLIVAE
MAV_4365|M.avium_104 YFVTDFDAQQAVLDDPVILLHQEKISSLPDLLPMLEKVAESGKPLLIIAE
Mflv_4924|M.gilvum_PYR-GCK YFVTDFDSQEAVLEDALVLLHREKISSLPDLLPLLEKVAEAGKPLLIVAE
Mvan_1495|M.vanbaalenii_PYR-1 YFVTDFDSQEAVLEDALVLLHREKISSLPDLLPLLEKVAEAGKPLLIIAE
MSMEG_1583|M.smegmatis_MC2_155 YFVTDFDSQEAVLEDALVLLHRDKISSLPDLLPLLEKVAEAGKPLLIVAE
TH_1687|M.thermoresistible__bu YFVTDFDTQQAVLEDAYVLLHREKISSLPDLLPLLEKVAESGKPLLVIAE
MAB_3731c|M.abscessus_ATCC_199 YFVTDFDDQKAVLEDALVLLHRDKISSLPDLLPLLELVAKEGKPLLIVAE
******* *:***:*. :***::******:***:** *: ****:::**
Mb3451c|M.bovis_AF2122/97 DVEGEALATLVVNAIRKTLKAVAVKGPYFGDRRKAFLEDLAVVTGGQVVN
Rv3417c|M.tuberculosis_H37Rv DVEGEALATLVVNAIRKTLKAVAVKGPYFGDRRKAFLEDLAVVTGGQVVN
MMAR_1126|M.marinum_M DIEGEALATLVVNSIRKTLKAIAVKSPYFGDRRKAFLQDLAAVTGAEVVN
MUL_0886|M.ulcerans_Agy99 DIEGEALATLVVNSIRKTLKAIAVKSPYFGDRRKAFLQDLAAVTGAEVVN
MLBr_00381|M.leprae_Br4923 DLEGEALATLVVNSIRKTLKAVAVKSPFFGDRRKAFLEDLAIVTGGQVVN
MAV_4365|M.avium_104 DIEGEALATLVVNSIRKTLKAVAVKAPFFGDRRKAFLEDLAIVTGGQVIN
Mflv_4924|M.gilvum_PYR-GCK DVEGEALSTLVVNAIRKTLKAVAVKAPFFGDRRKAFLDDLAVVTGGQVVN
Mvan_1495|M.vanbaalenii_PYR-1 DVEGEALSTLVVNAIRKTLKAVAVKAPFFGDRRKAFLDDLAVVTGGQVVN
MSMEG_1583|M.smegmatis_MC2_155 DVEGEALSTLVVNAIRKTLKAVAVKAPFFGDRRKAFLDDLAIVTGGQVVN
TH_1687|M.thermoresistible__bu DVEGEALSTLVVNAIRKTLKAVAVKAPFFGDRRKAFLEDLAIVTGGQVVN
MAB_3731c|M.abscessus_ATCC_199 DVEGEPLSTLVVNAIRKTLKAVAVKAPFFGDRRKAFLEDLAIVTNAQVVN
*:***.*:*****:*******:***.*:*********:*** **..:*:*
Mb3451c|M.bovis_AF2122/97 PDAGMVLREVGLEVLGSARRVVVSKDDTVIVDGGGTAEAVANRAKHLRAE
Rv3417c|M.tuberculosis_H37Rv PDAGMVLREVGLEVLGSARRVVVSKDDTVIVDGGGTAEAVANRAKHLRAE
MMAR_1126|M.marinum_M PDAGLVLREVGLEVMGSARRVVVSKDDTIIVDGGGAPEAVEARVNLLRSE
MUL_0886|M.ulcerans_Agy99 PDAGLVLREVGLEVMGSARRVVVSKDDTIIVDGGGAPEAVEARVNLLRSE
MLBr_00381|M.leprae_Br4923 PETGLVLREVGTDVLGSARRVVVSKDDTIIVDGGGSNDAVAKRVNQLRAE
MAV_4365|M.avium_104 PDTGLLLREVGTEVLGSARRVVVSKDDTIIVDGGGAKDAVANRIKQLRAE
Mflv_4924|M.gilvum_PYR-GCK PDVGLVLREVGLEVLGTARRVVVDKDSTVIVDGGGTQDAIEGRKGQLRAE
Mvan_1495|M.vanbaalenii_PYR-1 PDVGLVLRDVGLDVLGTARRVVVDKDSTVIVDGGGTQEAIEGRKAQLRAE
MSMEG_1583|M.smegmatis_MC2_155 PDVGLLLREVGLEVLGSARRVVVNKDSTVIVDGGGTAEAIADRVKQIKSE
TH_1687|M.thermoresistible__bu PDVGLVLREVGLEVLGTARRVVVTKDDTVIVDGGGTKEAIAERAAQLRKE
MAB_3731c|M.abscessus_ATCC_199 PDVGLSLREVGIEVLGTARRVVVSKDETVIVDGGGSEDAIKARVAQLKAE
*:.*: **:** :*:*:****** **.*:******: :*: * :: *
Mb3451c|M.bovis_AF2122/97 IDKSDSDWDREKLGERLAKLAGGVAVIKVGAATETALKERKESVEDAVAA
Rv3417c|M.tuberculosis_H37Rv IDKSDSDWDREKLGERLAKLAGGVAVIKVGAATETALKERKESVEDAVAA
MMAR_1126|M.marinum_M IDRSDSEWDREKLGERLAKLAGGVAVIKVGAATETELKKRKESVEDAVAA
MUL_0886|M.ulcerans_Agy99 IDRSDSEWDREKLGERLAKLAGGVAVIKVGAATETELKKRKESVEDAVAA
MLBr_00381|M.leprae_Br4923 IEVSDSEWDREKLQERVAKLAGGVAVIKVGAVTETALKKRKESVEDAVAA
MAV_4365|M.avium_104 IEKTDSDWDREKLQERLAKLAGGVAVIKVGAATETALKERKESVEDAVAA
Mflv_4924|M.gilvum_PYR-GCK IEVSDSDWDREKLEERLAKLAGGVAVIKVGAATETDLKKRKEAVEDAVAA
Mvan_1495|M.vanbaalenii_PYR-1 IEASDSDWDREKLEERLAKLAGGVAVIKVGAATETDLKKRKEAVEDAVHA
MSMEG_1583|M.smegmatis_MC2_155 IETTDSDWDREKLQERLAKLAGGVAVIKVGAATETDLKKRKEAVEDAVAA
TH_1687|M.thermoresistible__bu IETTDSDWDREKLEERLAKLAGGVAVIKVGAATETALKERKESVEDAVAA
MAB_3731c|M.abscessus_ATCC_199 IETTDSDWDREKLQERLAKLAGGVAVIKVGAPTDTALKERKHRVEDAVSA
*: :**:****** **:************** *:* **:**. ***** *
Mb3451c|M.bovis_AF2122/97 AKAAVEEGIVPGGGASLIHQARKALTELRASLTGDEVLGVDVFSEALAAP
Rv3417c|M.tuberculosis_H37Rv AKAAVEEGIVPGGGASLIHQARKALTELRASLTGDEVLGVDVFSEALAAP
MMAR_1126|M.marinum_M AKAAVEEGIVAGGGSALI-QARNALKDLRASLSGDEAVGVDVFSEALAAP
MUL_0886|M.ulcerans_Agy99 AKAAVEEGIVAGGGSALI-QARNALKDLRASLSGDEAVGVDVFSEALAAP
MLBr_00381|M.leprae_Br4923 AKASIEEGIIAGGGSALV-QCGAALKQLRTSLTGDEALGIDVFFEALKAP
MAV_4365|M.avium_104 AKAAVEEGIVAGGGSALL-QARKALDELRGSLSGDQALGVDVFAEALGAP
Mflv_4924|M.gilvum_PYR-GCK AKAAVEEGIVIGGGAALV-HAGSALDSLRSELKGDELLGVEVFASALSSP
Mvan_1495|M.vanbaalenii_PYR-1 AKAAVEEGIVVGGGAALV-QAGAGLAKLRSELSGDEALGVEVFTAALSSP
MSMEG_1583|M.smegmatis_MC2_155 AKAAVEEGIVTGGGAALV-QARSAVEKLRGELSGDEALGVDVFASALSAP
TH_1687|M.thermoresistible__bu AKAAVEEGVVVGGGVALL-QVRPALDKLRDSLSGDEALGVSIFAEALSAP
MAB_3731c|M.abscessus_ATCC_199 AKAAVEEGIVAGGGSALV-QARSALDGLRAELKGDEAKGVDVFEAALSAP
***::***:: *** :*: : .: ** .*.**: *:.:* ** :*
Mb3451c|M.bovis_AF2122/97 LFWIAANAGLDGSVVVNKVSELPAGHGLNVNTLSYGDLAADGVIDPVKVT
Rv3417c|M.tuberculosis_H37Rv LFWIAANAGLDGSVVVNKVSELPAGHGLNVNTLSYGDLAADGVIDPVKVT
MMAR_1126|M.marinum_M LYWIATNAGLDGSVVVNKVSELPAGHGLNAATLTYGDLAADGIVDPVKVT
MUL_0886|M.ulcerans_Agy99 LYWIATNAGLDGSVVVNKVSEVPAGHGLNAATLTYGDLAADGIVDPVKVT
MLBr_00381|M.leprae_Br4923 LYWIATNAGLDGAVVVDKVSGLPAGHGLNASTLGYGDLVADGVVDPVKVT
MAV_4365|M.avium_104 LYWIASNAGLDGAVAVHKVAELPAGHGLNAEKLSYGDLIADGVIDPVKVT
Mflv_4924|M.gilvum_PYR-GCK LYWIATNAGLDGAVVVNKVSELPAGQGFNAATLEYGDLIGDGVIDPVKVT
Mvan_1495|M.vanbaalenii_PYR-1 LYWIATNAGLDGAVVVNKVSELPAGQGFNAATLEYGDLIADGVIDPVKVT
MSMEG_1583|M.smegmatis_MC2_155 LYWIATNAGLDGSVVVNKVSELPKGQGFNAATLEFGDLVSAGVVDPAKVT
TH_1687|M.thermoresistible__bu LYWIATNAGLDGSVVVAKVAEMPDNHGFNAATLSYGDLMADGVIDPAKVT
MAB_3731c|M.abscessus_ATCC_199 LFWIATNAGLDGAVVVSKVAELDAGHGFNAATLEYGDLIADGVIDPVKVT
*:***:******:*.* **: : .:*:*. .* :*** . *::**.***
Mb3451c|M.bovis_AF2122/97 RSAVLNASSVARMVLTTETVVVDKPAKAEDHDH-H-HGHAH----
Rv3417c|M.tuberculosis_H37Rv RSAVLNASSVARMVLTTETVVVDKPAKAEDHDH-H-HGHAH----
MMAR_1126|M.marinum_M RSAVLNASSVARMVLTTETAIVDKPAEPEDDGHGH-HGHAH----
MUL_0886|M.ulcerans_Agy99 RSAVLNASSVARMVLTTETAIVDKPAEPEDDGHGH-HGHAH----
MLBr_00381|M.leprae_Br4923 RSAVLNAASVARMMLTTETAVVDKPAKTEEHDH---HGHAH----
MAV_4365|M.avium_104 RSAVLNSASVARMVLTTETAVVDKPAEEADDHG---HGHHHH---
Mflv_4924|M.gilvum_PYR-GCK RSAVVNAASVARMVLTTETAVVEKPAEAEDDGHGHGHGHHHH---
Mvan_1495|M.vanbaalenii_PYR-1 RSAVVNAASVARMILTTETAVVEKPEESEDDGHGHGHGHHHGHAH
MSMEG_1583|M.smegmatis_MC2_155 RSAVLNAASVARMILTTETAVVDKPADEDEHGHGHHHGHAH----
TH_1687|M.thermoresistible__bu RTAVLNAASVARMVLTTETAVVEKPEEEQDD---HGHGHVH----
MAB_3731c|M.abscessus_ATCC_199 RSAVINAASAARMILTTETSIVDKPAEEADHGHGH-HGHAH----
*:**:*::*.***:***** :*:** . :. *** *