For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MAKTIAYDEEARRGLERGLNALADAVKVTLGPKGRNVVLEKKWGAPTITNDGVSIAKEIELEDPYEKIGA ELVKEVAKKTDDVAGDGTTTATVLAQALVKEGLRNVAAGANPLGLKRGIEKAVEKVTETLLKSAKEVETK EQIAATAGISAGDQSIGDLIAEAMDKVGNEGVITVEESNTFGLQLELTEGMRFDKGYISGYFVTDAERQE AVLEDPYILLVSSKVSTVKDLLPLLEKVIQAGKPLLIIAEDVEGEALSTLVVNKIRGTFKSVAVKAPGFG DRRKAMLQDMAILTGGQVVSEEVGLSLETADITLLGTARKVVVTKDETTIVEGAGDSDAIAGRVAQIRSE IENSDSDYDREKLQERLAKLAGGVAVIKAGAATEVELKERKHRIEDAVRNAKAAVEEGIVAGGGVALLQA APSLDELSLTGDEATGAAIVKVAVEAPLKQIAFNSGLEPGVVAEKVRNSPSGTGLNAATGEYEDLLAAGV ADPVKVTRSALQNAASIAALFLTTEAVVADKPEKAAAPAGDPTGGMGGMDF
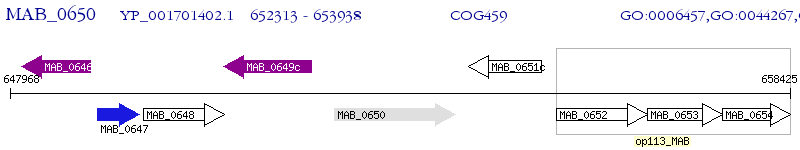
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_0650 | groEL | - | 100% (541) | chaperonin GroEL |
| M. abscessus ATCC 19977 | MAB_3731c | - | e-180 | 63.07% (528) | 60 kDa chaperonin 1 (GroEL protein 1) |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0448 | groEL | 0.0 | 93.74% (543) | chaperonin GroEL |
| M. gilvum PYR-GCK | Mflv_0141 | groEL | 0.0 | 94.82% (541) | chaperonin GroEL |
| M. tuberculosis H37Rv | Rv0440 | groEL | 0.0 | 93.74% (543) | chaperonin GroEL |
| M. leprae Br4923 | MLBr_00317 | groEL | 0.0 | 94.09% (541) | chaperonin GroEL |
| M. marinum M | MMAR_0759 | groEL | 0.0 | 96.49% (541) | 60 kDa chaperonin 2 GroEL2 |
| M. avium 104 | MAV_4707 | groEL | 0.0 | 96.12% (541) | chaperonin GroEL |
| M. smegmatis MC2 155 | MSMEG_0880 | groEL | 0.0 | 95.75% (541) | chaperonin GroEL |
| M. thermoresistible (build 8) | TH_0975 | groEL1 | 0.0 | 95.01% (541) | 60 KDA CHAPERONIN 1 GROEL1 (PROTEIN CPN60-1) (GROEL |
| M. ulcerans Agy99 | MUL_1393 | groEL | 0.0 | 96.30% (541) | chaperonin GroEL |
| M. vanbaalenii PYR-1 | Mvan_0764 | groEL | 0.0 | 95.75% (541) | chaperonin GroEL |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_0141|M.gilvum_PYR-GCK MAKTIAYDEEARRGLERGLNSLADAVKVTLGPKGRNVVLEKKWGAPTITN
Mvan_0764|M.vanbaalenii_PYR-1 MSKIIAYDEEARRGLERGLNALADAVKVTLGPKGRNVVLEKKWGAPTITN
MSMEG_0880|M.smegmatis_MC2_155 MAKTIAYDEEARRGLERGLNSLADAVKVTLGPKGRNVVLEKKWGAPTITN
MAB_0650|M.abscessus_ATCC_1997 MAKTIAYDEEARRGLERGLNALADAVKVTLGPKGRNVVLEKKWGAPTITN
TH_0975|M.thermoresistible__bu MAKQIAYDEEARRGLERGLNALADAVKVTLGPKGRNVVLEKKWGAPTITN
MMAR_0759|M.marinum_M MAKTIAYDEEARRGLERGLNALADAVKVTLGPKGRNVVLEKKWGAPTITN
MUL_1393|M.ulcerans_Agy99 MAKTIAYDEEARRGLERGLNALADAVKVTLGPKGRNVVLEKKWGAPTITN
MAV_4707|M.avium_104 MAKTIAYDEEARRGLERGLNALADAVKVTLGPKGRNVVLEKKWGAPTITN
Mb0448|M.bovis_AF2122/97 MAKTIAYDEEARRGLERGLNALADAVKVTLGPKGRNVVLEKKWGAPTITN
Rv0440|M.tuberculosis_H37Rv MAKTIAYDEEARRGLERGLNALADAVKVTLGPKGRNVVLEKKWGAPTITN
MLBr_00317|M.leprae_Br4923 MAKTIAYDEEARRGLERGLNSLADAVKVTLGPKGRNVVLEKKWGAPTITN
*:* ****************:*****************************
Mflv_0141|M.gilvum_PYR-GCK DGVSIAKEIELEDPYEKIGAELVKEVAKKTDDVAGDGTTTATVLAQALVR
Mvan_0764|M.vanbaalenii_PYR-1 DGVSIAKEIELEDPYEKIGAELVKEVAKKTDDVAGDGTTTATVLAQALVR
MSMEG_0880|M.smegmatis_MC2_155 DGVSIAKEIELEDPYEKIGAELVKEVAKKTDDVAGDGTTTATVLAQALVR
MAB_0650|M.abscessus_ATCC_1997 DGVSIAKEIELEDPYEKIGAELVKEVAKKTDDVAGDGTTTATVLAQALVK
TH_0975|M.thermoresistible__bu DGVSIAKEIELEDPYEKIGAELVKEVAKKTDDVAGDGTTTATVLAQALVK
MMAR_0759|M.marinum_M DGVSIAKEIELEDPYEKIGAELVKEVAKKTDDVAGDGTTTATVLAQALVK
MUL_1393|M.ulcerans_Agy99 DGVSIAKEIELEDPYEKIGAELVKEVAKKTDDVAGDGTTTATVLAQALVK
MAV_4707|M.avium_104 DGVSIAKEIELEDPYEKIGAELVKEVAKKTDDVAGDGTTTATVLAQALVR
Mb0448|M.bovis_AF2122/97 DGVSIAKEIELEDPYEKIGAELVKEVAKKTDDVAGDGTTTATVLAQALVR
Rv0440|M.tuberculosis_H37Rv DGVSIAKEIELEDPYEKIGAELVKEVAKKTDDVAGDGTTTATVLAQALVR
MLBr_00317|M.leprae_Br4923 DGVSIAKEIELEDPYEKIGAELVKEVAKKTDDVAGDGTTTATVLAQALVK
*************************************************:
Mflv_0141|M.gilvum_PYR-GCK EGLRNVAAGANPLGLKRGIEKAVEAVTQGLLKSAKEVETKEQIAATAAIS
Mvan_0764|M.vanbaalenii_PYR-1 EGLRNVAAGANPLGLKRGIEKAVEKVTETLLKSAKEVETKEQIAATAAIS
MSMEG_0880|M.smegmatis_MC2_155 EGLRNVAAGANPLGLKRGIEKAVEKVTETLLKSAKEVETKEQIAATAGIS
MAB_0650|M.abscessus_ATCC_1997 EGLRNVAAGANPLGLKRGIEKAVEKVTETLLKSAKEVETKEQIAATAGIS
TH_0975|M.thermoresistible__bu EGLRNVAAGANPLALKRGIEAAVEKVTETLLKSAKEVETKEQIANTAAIS
MMAR_0759|M.marinum_M EGLRNVAAGANPLGLKRGIEKAVEKVTETLLKSAKEVETKEQIAATAAIS
MUL_1393|M.ulcerans_Agy99 EGLRNVAAGANPLGLKRGIEKAVEKVTETLLKSAKEVETKEQIAATAAIS
MAV_4707|M.avium_104 EGLRNVAAGANPLGLKRGIEKAVEKVTETLLKSAKEVETKDQIAATAAIS
Mb0448|M.bovis_AF2122/97 EGLRNVAAGANPLGLKRGIEKAVEKVTETLLKGAKEVETKEQIAATAAIS
Rv0440|M.tuberculosis_H37Rv EGLRNVAAGANPLGLKRGIEKAVEKVTETLLKGAKEVETKEQIAATAAIS
MLBr_00317|M.leprae_Br4923 EGLRNVAAGANPLGLKRGIEKAVDKVTETLLKDAKEVETKEQIAATAAIS
*************.****** **: **: ***.*******:*** **.**
Mflv_0141|M.gilvum_PYR-GCK AGDVQIGELIAEAMDKVGNEGVITVEESNTFGLQLELTEGMRFDKGYISG
Mvan_0764|M.vanbaalenii_PYR-1 AGDTQIGELIAEAMDKVGNEGVITVEESNTFGLQLELTEGMRFDKGYISG
MSMEG_0880|M.smegmatis_MC2_155 AGDQSIGDLIAEAMDKVGNEGVITVEESNTFGLQLELTEGMRFDKGYISG
MAB_0650|M.abscessus_ATCC_1997 AGDQSIGDLIAEAMDKVGNEGVITVEESNTFGLQLELTEGMRFDKGYISG
TH_0975|M.thermoresistible__bu AGDQQIGELIAEAMDKVGNEGVITVEESQTFGLQLELTEGMRFDKGYISG
MMAR_0759|M.marinum_M AGDQSIGDLIAEAMDKVGNEGVITVEESNTFGLQLELTEGMRFDKGYISG
MUL_1393|M.ulcerans_Agy99 AGDQSIGDLIAEAMDKVGNEGVITVEESNTFGLQLELTEGMRFDKGYISG
MAV_4707|M.avium_104 AGDQSIGDLIAEAMDKVGNEGVITVEESNTFGLQLELTEGMRFDKGYISG
Mb0448|M.bovis_AF2122/97 AGDQSIGDLIAEAMDKVGNEGVITVEESNTFGLQLELTEGMRFDKGYISG
Rv0440|M.tuberculosis_H37Rv AGDQSIGDLIAEAMDKVGNEGVITVEESNTFGLQLELTEGMRFDKGYISG
MLBr_00317|M.leprae_Br4923 AGDQSIGDLIAEAMDKVGNEGVITVEESNTFGLQLELTEGMRFDKGYISG
*** .**:********************:*********************
Mflv_0141|M.gilvum_PYR-GCK YFVTDAERQEAVLEDPYILLVSSKVSTVKDLLPLLEKVIQAGKPLLIIAE
Mvan_0764|M.vanbaalenii_PYR-1 YFVTDAERQEAVLEDPYILLVSSKVSTVKDLLPLLEKVIQAGKPLLIIAE
MSMEG_0880|M.smegmatis_MC2_155 YFVTDAERQEAVLEDPYILLVSSKVSTVKDLLPLLEKVIQSGKPLLIIAE
MAB_0650|M.abscessus_ATCC_1997 YFVTDAERQEAVLEDPYILLVSSKVSTVKDLLPLLEKVIQAGKPLLIIAE
TH_0975|M.thermoresistible__bu YFVTDAERQEAVLEDPYILLVSSKVSTVKDLLPLLEKVIQSGKPLLIIAE
MMAR_0759|M.marinum_M YFVTDAERQEAVLEDPYILLVSSKVSTVKDLLPLLEKVIQGGKPLLIIAE
MUL_1393|M.ulcerans_Agy99 YFVTDAERQEAVLEDPYILLVSSKVSTVKDLLPLLEKVIQGGKPLLIIAE
MAV_4707|M.avium_104 YFVTDAERQEAVLEDPFILLVSSKVSTVKDLLPLLEKVIQAGKPLLIIAE
Mb0448|M.bovis_AF2122/97 YFVTDPERQEAVLEDPYILLVSSKVSTVKDLLPLLEKVIGAGKPLLIIAE
Rv0440|M.tuberculosis_H37Rv YFVTDPERQEAVLEDPYILLVSSKVSTVKDLLPLLEKVIGAGKPLLIIAE
MLBr_00317|M.leprae_Br4923 YFVTDAERQEAVLEEPYILLVSSKVSTVKDLLPLLEKVIQAGKSLLIIAE
*****.********:*:********************** .**.******
Mflv_0141|M.gilvum_PYR-GCK DVEGEALSTLVVNKIRGTFKSVAVKAPGFGDRRKAMLQDMAILTGGQVVS
Mvan_0764|M.vanbaalenii_PYR-1 DVEGEALSTLVVNKIRGTFKSVAVKAPGFGDRRKAMLQDIAILTGGQVVS
MSMEG_0880|M.smegmatis_MC2_155 DVEGEALSTLVVNKIRGTFKSVAVKAPGFGDRRKAMLQDMAILTGGQVIS
MAB_0650|M.abscessus_ATCC_1997 DVEGEALSTLVVNKIRGTFKSVAVKAPGFGDRRKAMLQDMAILTGGQVVS
TH_0975|M.thermoresistible__bu DVEGEALSTLVVNKIRGTFKSVAVKAPGFGDRRKAMLQDMAILTGGQVIS
MMAR_0759|M.marinum_M DVEGEALSTLVVNKIRGTFKSVAVKAPGFGDRRKAMLQDMAILTGGQVIS
MUL_1393|M.ulcerans_Agy99 DVEGEALSTLVVNKIRGTFKSVAVKAPGFGDRRKAMLQDMAILTGGQVIS
MAV_4707|M.avium_104 DVEGEALSTLVVNKIRGTFKSVAVKAPGFGDRRKAMLQDMAILTGGQVIS
Mb0448|M.bovis_AF2122/97 DVEGEALSTLVVNKIRGTFKSVAVKAPGFGDRRKAMLQDMAILTGGQVIS
Rv0440|M.tuberculosis_H37Rv DVEGEALSTLVVNKIRGTFKSVAVKAPGFGDRRKAMLQDMAILTGGQVIS
MLBr_00317|M.leprae_Br4923 DVEGEALSTLVVNKIRGTFKSVAVKAPGFGDRRKAMLQDMAILTGAQVIS
***************************************:*****.**:*
Mflv_0141|M.gilvum_PYR-GCK EEVGLSLETADVALLGTARKVVVTKDETTIVEGAGDSDAIAGRVAQIRAE
Mvan_0764|M.vanbaalenii_PYR-1 EEVGLSLETADVALLGTARKVVVTKDETTIVEGAGDSDAIAGRVAQIRAE
MSMEG_0880|M.smegmatis_MC2_155 EEVGLSLETADVSLLGKARKVVVTKDETTIVEGAGDAEAIQGRVAQIRAE
MAB_0650|M.abscessus_ATCC_1997 EEVGLSLETADITLLGTARKVVVTKDETTIVEGAGDSDAIAGRVAQIRSE
TH_0975|M.thermoresistible__bu EEVGLSLETADISLLGQARKVVVTKDETTIVEGAGDPDAIAGRVAQIRQE
MMAR_0759|M.marinum_M EEVGLSLETADISLLGKARKVVITKDETTIVEGAGDSDAIAGRVAQIRAE
MUL_1393|M.ulcerans_Agy99 EEVGLSLETADISLLGKARKVVITKDETTLVEGAGDSDAIAGRVAQIRAE
MAV_4707|M.avium_104 EEVGLSLESADISLLGKARKVVVTKDETTIVEGAGDSDAIAGRVAQIRTE
Mb0448|M.bovis_AF2122/97 EEVGLTLENADLSLLGKARKVVVTKDETTIVEGAGDTDAIAGRVAQIRQE
Rv0440|M.tuberculosis_H37Rv EEVGLTLENADLSLLGKARKVVVTKDETTIVEGAGDTDAIAGRVAQIRQE
MLBr_00317|M.leprae_Br4923 EEVGLTLENTDLSLLGKARKVVMTKDETTIVEGAGDTDAIAGRVAQIRTE
*****:**.:*::*** *****:******:******.:** ******* *
Mflv_0141|M.gilvum_PYR-GCK IENSDSDYDREKLQERLAKLAGGVAVIKAGAATEVELKERKHRIEDAVRN
Mvan_0764|M.vanbaalenii_PYR-1 IENSDSDYDREKLQERLAKLAGGVAVIKAGAATEVELKERKHRIEDAVRN
MSMEG_0880|M.smegmatis_MC2_155 IENSDSDYDREKLQERLAKLAGGVAVIKAGAATEVELKERKHRIEDAVRN
MAB_0650|M.abscessus_ATCC_1997 IENSDSDYDREKLQERLAKLAGGVAVIKAGAATEVELKERKHRIEDAVRN
TH_0975|M.thermoresistible__bu IENSDSDYDREKLQERLAKLAGGVAVIKAGAATEVELKERKHRIEDAVRN
MMAR_0759|M.marinum_M IENSDSDYDREKLQERLAKLAGGVAVIKAGAATEVELKERKHRIEDAVRN
MUL_1393|M.ulcerans_Agy99 IENSDSDYDREKLQERLAKLAGGVAVIKAGAATEVELKERKHRIEDAVRN
MAV_4707|M.avium_104 IENSDSDYDREKLQERLAKLAGGVAVIKAGAATEVELKERKHRIEDAVRN
Mb0448|M.bovis_AF2122/97 IENSDSDYDREKLQERLAKLAGGVAVIKAGAATEVELKERKHRIEDAVRN
Rv0440|M.tuberculosis_H37Rv IENSDSDYDREKLQERLAKLAGGVAVIKAGAATEVELKERKHRIEDAVRN
MLBr_00317|M.leprae_Br4923 IENSDSDYDREKLQERLAKLAGGVAVIKAGAATEVELKERKHRIEDAVRN
**************************************************
Mflv_0141|M.gilvum_PYR-GCK AKAAVEEGIVAGGGVALLQSAPVLEDLGLSGDEATGANIVRVALSAPLKQ
Mvan_0764|M.vanbaalenii_PYR-1 AKAAVEEGIVAGGGVALLQSAPSLDELNLTGDEATGANIVRVALSAPLKQ
MSMEG_0880|M.smegmatis_MC2_155 AKAAVEEGIVAGGGVALLQSAPSLEELSLTGDEATGANIVRVALSAPLKQ
MAB_0650|M.abscessus_ATCC_1997 AKAAVEEGIVAGGGVALLQAAPSLDELSLTGDEATGAAIVKVAVEAPLKQ
TH_0975|M.thermoresistible__bu AKAAVEEGIVAGGGVSLLQAAPVLDELKLEGDEATGVNIVRVALEAPLKQ
MMAR_0759|M.marinum_M AKAAVEEGIVAGGGVALLHAVPSLDELKLSGDEATGANIVRVALEAPLKQ
MUL_1393|M.ulcerans_Agy99 AKAAVEEGIVAGGGVALLHAVPSLDELKLSGDEATGANIVRVALEAPLKQ
MAV_4707|M.avium_104 AKAAVEEGIVAGGGVALLHAIPALDELKLEGDEATGANIVRVALEAPLKQ
Mb0448|M.bovis_AF2122/97 AKAAVEEGIVAGGGVTLLQAAPTLDELKLEGDEATGANIVKVALEAPLKQ
Rv0440|M.tuberculosis_H37Rv AKAAVEEGIVAGGGVTLLQAAPTLDELKLEGDEATGANIVKVALEAPLKQ
MLBr_00317|M.leprae_Br4923 AKAAVEEGIVAGGGVTLLQAAPALDKLKLTGDEATGANIVKVALEAPLKQ
***************:**:: * *:.* * ******. **:**:.*****
Mflv_0141|M.gilvum_PYR-GCK IAFNGGLEPGVVAEKVSNLPAGHGLNAATGEYEDLLKAGVADPVKVTRSA
Mvan_0764|M.vanbaalenii_PYR-1 IAFNGGLEPGVVAEKVTNLPAGHGLNAATGEYEDLLKAGVADPVKVTRSA
MSMEG_0880|M.smegmatis_MC2_155 IALNGGLEPGVVAEKVSNLPAGHGLNAATGEYEDLLAAGVADPVKVTRSA
MAB_0650|M.abscessus_ATCC_1997 IAFNSGLEPGVVAEKVRNSPSGTGLNAATGEYEDLLAAGVADPVKVTRSA
TH_0975|M.thermoresistible__bu IAANSGLEPGVVAEKVRNLEAGKGLNAATGEYEDLLAAGVADPVKVTRSA
MMAR_0759|M.marinum_M IAFNGGLEPGVVAEKVRNSPAGTGLNAATGVYEDLLKAGVADPVKVTRSA
MUL_1393|M.ulcerans_Agy99 IAFNGGLEPGVVAEKVRNSPVGTGLNAATGVYEDLLKAGVADPVKVTRSA
MAV_4707|M.avium_104 IAFNGGLEPGVVAEKVRNSPAGTGLNAATGEYEDLLKAGVADPVKVTRSA
Mb0448|M.bovis_AF2122/97 IAFNSGLEPGVVAEKVRNLPAGHGLNAQTGVYEDLLAAGVADPVKVTRSA
Rv0440|M.tuberculosis_H37Rv IAFNSGLEPGVVAEKVRNLPAGHGLNAQTGVYEDLLAAGVADPVKVTRSA
MLBr_00317|M.leprae_Br4923 IAFNSGMEPGVVAEKVRNLSVGHGLNAATGEYEDLLKAGVADPVKVTRSA
** *.*:********* * * **** ** ***** *************
Mflv_0141|M.gilvum_PYR-GCK LQNAASIAALFLTTEAVVADKPEKAAAPAGDPTGGMGGMDF
Mvan_0764|M.vanbaalenii_PYR-1 LQNAASIAALFLTTEAVVADKPEKAAAPAGDPTGGMGGMDF
MSMEG_0880|M.smegmatis_MC2_155 LQNAASIAALFLTTEAVVADKPEKAAAPAGDPTGGMGGMDF
MAB_0650|M.abscessus_ATCC_1997 LQNAASIAALFLTTEAVVADKPEKAAAPAGDPTGGMGGMDF
TH_0975|M.thermoresistible__bu LQNAASIAALFLTTEAVVADKPEKAAAPAGDPTGGMGGMDF
MMAR_0759|M.marinum_M LQNAASIAGLFLTTEAVVADKPEKAAAPAGDPTGGMGGMDF
MUL_1393|M.ulcerans_Agy99 LQNAASIAGLFLTTEAVVADKPEKAAAPAGDPTGGMGGMDF
MAV_4707|M.avium_104 LQNAASIAGLFLTTEAVVADKPEKAAAPAGDPTGGMGGMDF
Mb0448|M.bovis_AF2122/97 LQNAASIAGLFLTTEAVVADKPEKEKASVPG-GGDMGGMDF
Rv0440|M.tuberculosis_H37Rv LQNAASIAGLFLTTEAVVADKPEKEKASVPG-GGDMGGMDF
MLBr_00317|M.leprae_Br4923 LQNAASIAGLFLTTEAVVADKPEKTAAPASDPTGGMGGMDF
********.*************** *.. . *.******