For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SGTVKADAVILVGGQGTRLRPLTLSAPKPMLPIAGFPFLTHVLSRVAAAGIDHVVLGTSYKADVFESEFG DGSKLGLQITYVYEEEPLGTGGAIRNVLEHLRYDTALIFNGDVLSGLDLKDLLAQHEQTQADLTLHLVRV GDPRAFGCVPTDSDGKVTAFLEKTEDPPTDQINAGCYVFRRELIEQIPSGRPVSVEREVFPGLLSSGAKV CGYVDTSYWRDMGTPEDFVRGSADLVRGIAPSPAIPEHPGEALVHDGASVAPGALVIGGTVVGRGAEIGP GARLDGAVVFDGARIEAGAVVERSIIGFGARIGPRALVRDGVIGDGADIGARCELLRGARVWPGITIPDG GIRYSSDV*
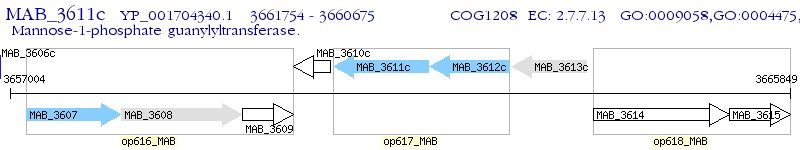
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_3611c | - | - | 100% (359) | putative sugar-phosphate nucleotidyl transferase |
| M. abscessus ATCC 19977 | MAB_1352 | glgC | 4e-23 | 27.61% (373) | glucose-1-phosphate adenylyltransferase |
| M. abscessus ATCC 19977 | MAB_4113 | - | 9e-19 | 32.40% (179) | glucose-1-phosphate thymidylyltransferase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3292c | manC | 1e-165 | 78.71% (357) | mannose-1-phosphate guanyltransferase |
| M. gilvum PYR-GCK | Mflv_4734 | - | 1e-169 | 80.22% (359) | nucleotidyl transferase |
| M. tuberculosis H37Rv | Rv3264c | manB | 1e-165 | 78.71% (357) | D-alpha-D-mannose-1-phosphate guanylyltransferase MANB |
| M. leprae Br4923 | MLBr_00753 | rmlA2 | 1e-165 | 78.15% (357) | putative sugar-phosphate nucleotidyl transferase |
| M. marinum M | MMAR_1277 | manB | 1e-165 | 79.89% (353) | d-alpha-D-mannose-1-phosphate guanylyltransferase ManB |
| M. avium 104 | MAV_4228 | - | 1e-164 | 78.21% (358) | nucleotidyl transferase |
| M. smegmatis MC2 155 | MSMEG_5078 | glgC | 9e-24 | 27.61% (373) | glucose-1-phosphate adenylyltransferase |
| M. thermoresistible (build 8) | TH_0779 | rmlA2 | 1e-171 | 79.94% (359) | D-ALPHA-D-MANNOSE-1-PHOSPHATE GUANYLYLTRANSFERASE MANB |
| M. ulcerans Agy99 | MUL_2610 | manB | 1e-166 | 79.89% (353) | d-alpha-D-mannose-1-phosphate guanylyltransferase ManB |
| M. vanbaalenii PYR-1 | Mvan_1729 | - | 1e-171 | 81.36% (354) | nucleotidyl transferase |
CLUSTAL 2.0.9 multiple sequence alignment
MLBr_00753|M.leprae_Br4923 -MATLQVDAVVLVGGKGTRLRPLTLSAPKPMLPTAG-LPFLTHLLSRIAA
MAV_4228|M.avium_104 -MGTPEVDVVILVGGKGTRLRPLTLSAPKPMLPTAG-LPFLTHLLSRIAA
MMAR_1277|M.marinum_M -MATNGVDAVVLVGGKGTRLRPLTLSAPKPMLPTAG-LPFLTHLLSRIAA
MUL_2610|M.ulcerans_Agy99 -MATNGVDAVVLVGGKGTRLRPLTLSAPKPMLPTAG-LPFLTHLLSRIAA
Mb3292c|M.bovis_AF2122/97 -MATHQVDAVVLVGGKGTRLRPLTLSAPKPMLPTAG-LPFLTHLLSRIAA
Rv3264c|M.tuberculosis_H37Rv -MATHQVDAVVLVGGKGTRLRPLTLSAPKPMLPTAG-LPFLTHLLSRIAA
TH_0779|M.thermoresistible__bu MVNPAQVDAVVLVGGQGTRLRPLTLSAPKPMLPTAG-LPFLTHLLSRIAE
Mflv_4734|M.gilvum_PYR-GCK MVNPAQVDAVVLVGGMGTRLRPLTLSAPKPMLPTAG-LPFLTHLLSRIAA
Mvan_1729|M.vanbaalenii_PYR-1 MVNPAEVDAVVLVGGMGTRLRPLTLSAPKPMLPTAG-LPFLTHLLSRIAD
MAB_3611c|M.abscessus_ATCC_199 MSGTVKADAVILVGGQGTRLRPLTLSAPKPMLPIAG-FPFLTHVLSRVAA
MSMEG_5078|M.smegmatis_MC2_155 MRELPHVLGIVLAGGEGKRLYPLTADRAKPAVPFGGAYRLIDFVLSNLVN
. ::*.** *.** *** . .** :* .* :: .:**.:.
MLBr_00753|M.leprae_Br4923 AGIEHVILSTSYRDAVFEAEFGDGSKLG-LQIDYVIEESPLGTGG-----
MAV_4228|M.avium_104 AGIEHVILSTSYQAAVFEAEFGDGSKLG-LQIEYVTEERPLGTGG-----
MMAR_1277|M.marinum_M AGIEHVVLSTSYQAGVFEAEFGDGSKLG-LQIDYVTEDKPLGTGG-----
MUL_2610|M.ulcerans_Agy99 AGIEHVVLSTSYQAGVFEAEFGDGSKLG-LQIDYVTEDKPLGTGG-----
Mb3292c|M.bovis_AF2122/97 AGIEHVILGTSYKPAVFEAEFGDGSALG-LQIEYVTEEHPLGTGG-----
Rv3264c|M.tuberculosis_H37Rv AGIEHVILGTSYKPAVFEAEFGDGSALG-LQIEYVTEEHPLGTGG-----
TH_0779|M.thermoresistible__bu AGIEHVVLGTSYKAHVFEAEFGDGSKFG-LNIEYVTETEPLGTGG-----
Mflv_4734|M.gilvum_PYR-GCK AGIEHVVMGTSYKAAVFESEFGDGSKLG-LQIDYVVEDEALGTGG-----
Mvan_1729|M.vanbaalenii_PYR-1 AGIEHVVLGTSYKANVFESEFGDGSKLG-LQIDYVVEDEPLGTGG-----
MAB_3611c|M.abscessus_ATCC_199 AGIDHVVLGTSYKADVFESEFGDGSKLG-LQITYVYEEEPLGTGG-----
MSMEG_5078|M.smegmatis_MC2_155 ARYLRICVLTQYKSHSLDRHISQNWRLSGLAGEYITPVPAQQRLGPRWYT
* :: : *.*: :: .:.:. :. * *: . *
MLBr_00753|M.leprae_Br4923 -------GIANVIDQLRHDTVMVFNGDVLSGVDLGQLLGFQRTNFADVTL
MAV_4228|M.avium_104 -------GIANVAGQLRHDTVMVFNGDVLSGADLGQMLDFHAAQQADVTL
MMAR_1277|M.marinum_M -------GIVNVAGKLHHDTVMVFNGDVLSGADLGQLLDYHHQNQADVTL
MUL_2610|M.ulcerans_Agy99 -------GIVNVAGKLHHDTVMVFNGDVLSGADLGQLLDYHHQNQADVTL
Mb3292c|M.bovis_AF2122/97 -------GIANVAGKLRNDTAMVFNGDVLSGADLAQLLDFHRSNRADVTL
Rv3264c|M.tuberculosis_H37Rv -------GIANVAGKLRNDTAMVFNGDVLSGADLAQLLDFHRSNRADVTL
TH_0779|M.thermoresistible__bu -------AIANVADKLRFDTVMVFNGDVLSGADLQGLLDTHERSGADLTL
Mflv_4734|M.gilvum_PYR-GCK -------GIANVSSKLRHDTAVVFNGDVLSGCDLGALLDSHEERGADVTL
Mvan_1729|M.vanbaalenii_PYR-1 -------GIANVASKLRYDTAVVFNGDVLSGCDLRALLDSHVSRDADVTL
MAB_3611c|M.abscessus_ATCC_199 -------AIRNVLEHLRYDTALIFNGDVLSGLDLKDLLAQHEQTQADLTL
MSMEG_5078|M.smegmatis_MC2_155 GSADAIYQSLNLIYDEDPDYIIIFGADHVYRMDPEQMMQFHIESGAGATV
*: . * ::*..* : * :: : *. *:
MLBr_00753|M.leprae_Br4923 HLVRVG--DPRAFGCVST-EDGRVTAFLEKTQDPP-------TDQINAGC
MAV_4228|M.avium_104 HLVRVS--DPRAFGCVTT-EDGRVTAFLEKTQDPP-------TDQINAGC
MMAR_1277|M.marinum_M HLVRVG--DPRAFGCVPT-EDGRVTAFLEKTQDPP-------TDQINAGC
MUL_2610|M.ulcerans_Agy99 HLVRVG--DPRAFGCVPT-EDGRVTAFLEKTQDPP-------TDQINAGC
Mb3292c|M.bovis_AF2122/97 QLVRVG--DPRAFGCVPTDEEDRVVAFLEKTEDPP-------TDQINAGC
Rv3264c|M.tuberculosis_H37Rv QLVRVG--DPRAFGCVPTDEEDRVVAFLEKTEDPP-------TDQINAGC
TH_0779|M.thermoresistible__bu HLVRVG--DPRAFGCVPTDAHGVVTAFLEKTQDPP-------TDQINAGC
Mflv_4734|M.gilvum_PYR-GCK HLVRVG--DPRAFGCVPTDSDGVVTAFLEKTQDPP-------TDQINAGC
Mvan_1729|M.vanbaalenii_PYR-1 HLVRVG--DPRAFGCVPTDSDGNVTAFLEKTQDPP-------TDQINAGC
MAB_3611c|M.abscessus_ATCC_199 HLVRVG--DPRAFGCVPTDSDGKVTAFLEKTEDPP-------TDQINAGC
MSMEG_5078|M.smegmatis_MC2_155 AGIRVPRSEASAFGCIDADESGRIREFIEKPADPPGTPDDPEQTFVSMGN
:** :. ****: : . : *:**. *** :. *
MLBr_00753|M.leprae_Br4923 YVFERNVIDRIPRG------REVSVEREVFPTLLSDADVKVCGY------
MAV_4228|M.avium_104 YVFARRVIDRIPRG------REVSVEREVFPALLSDPDVTVCGY------
MMAR_1277|M.marinum_M YVFKREIIDRIPRG------REVSVEREVFPSLLSDPDIKIYGY------
MUL_2610|M.ulcerans_Agy99 YVFKREIIDRIPRG------REVSVEREVFPSLLSDPDIKIYGY------
Mb3292c|M.bovis_AF2122/97 YVFERNVIDRIPQG------REVSVEREVFPALLADGDCKIYGY------
Rv3264c|M.tuberculosis_H37Rv YVFERNVIDRIPQG------REVSVEREVFPALLADGDCKIYGY------
TH_0779|M.thermoresistible__bu YVFQRHVIDRIPRG------RAVSVEREVFPQLLSEG-LRICGH------
Mflv_4734|M.gilvum_PYR-GCK YVFKRSIIEAIPKG------RALSVEREVFPKLLSDG-KKVCGY------
Mvan_1729|M.vanbaalenii_PYR-1 YVFKRSVIDAIPKG------RALSVEREIFPQLLSDG-KRVCGY------
MAB_3611c|M.abscessus_ATCC_199 YVFRRELIEQIPSG------RPVSVEREVFPGLLSSG-AKVCGY------
MSMEG_5078|M.smegmatis_MC2_155 YIFTTKVLIDAIRADADDDHSDHDMGGDIIPRLVADGMAAVYDFKNNEVP
*:* :: . .: :::* *::. : ..
MLBr_00753|M.leprae_Br4923 ----VDATYWRDMGTPEDFVRGSADLVRGIAPSPALGGHRGEHLVHDGAA
MAV_4228|M.avium_104 ----VDATYWRDMGTPEDFVRGSADLVRGIAPSPALHGHRGEQLVHDGAA
MMAR_1277|M.marinum_M ----VDATYWRDMGTPEDFVRGSADLVRGIAPSPALQGQRGEQLVHDGAA
MUL_2610|M.ulcerans_Agy99 ----VDATYWRDMGTPEDFVRGSADLVRGIAPSPALQGQRGEQLVHDGAA
Mb3292c|M.bovis_AF2122/97 ----VDASYWRDMGTPEDFVRGSADLVRGIAPSPALRGHRGEQLVHDGAA
Rv3264c|M.tuberculosis_H37Rv ----VDASYWRDMGTPEDFVRGSADLVRGIAPSPALRGHRGEQLVHDGAA
TH_0779|M.thermoresistible__bu ----VDASYWRDMGTPEDFVRGSADLVRGIAPSPALRGQRGESLVHDGAA
Mflv_4734|M.gilvum_PYR-GCK ----VDATYWRDMGTPEDFVRGSADLVRGIAPSPALTHQRGEKLVHDGAS
Mvan_1729|M.vanbaalenii_PYR-1 ----VDATYWRDMGTPEDFVRGSADLVRGIAPSPALHGHRGESLVHDGAS
MAB_3611c|M.abscessus_ATCC_199 ----VDTSYWRDMGTPEDFVRGSADLVRGIAPSPAIPEHPGEALVHDGAS
MSMEG_5078|M.smegmatis_MC2_155 GATERDHGYWRDVGTLDAFYDAHMDLVS-VHPVFNLYNKR-WPIRGESEN
* ****:** : * . *** : * : : : :.
MLBr_00753|M.leprae_Br4923 VSPGAVLIGG----TVVGRGAEIGPGVRLDGAVIFDGAKVEAGSVIERSI
MAV_4228|M.avium_104 VSPGAVLIGG----TVVGRGAEIGPGVRLDGAVIFDGVKVEAGSVIERSI
MMAR_1277|M.marinum_M IAPGAVLIGG----TVVGRGAEIGPGVRLDGAVIFDGVKVEAGSVIERSI
MUL_2610|M.ulcerans_Agy99 IAPGAVLIGG----TVVGRGAEIGPGVRLDGAVIFDGVKVEAGSVIERSI
Mb3292c|M.bovis_AF2122/97 VSPGALLIGG----TVVGRGAEIGPGTRLDGAVIFDGVRVEAGCVIERSI
Rv3264c|M.tuberculosis_H37Rv VSPGALLIGG----TVVGRGAEIGPGTRLDGAVIFDGVRVEAGCVIERSI
TH_0779|M.thermoresistible__bu VAPGALLIGG----TVVGRGAEIGPGARLDGAVIFDGVRVEAGSVIERSI
Mflv_4734|M.gilvum_PYR-GCK VAPGALLIGG----TVVGRGAEVAGGARLDGAVIFDGVKIGAGAVIERSI
Mvan_1729|M.vanbaalenii_PYR-1 VAPGALLIGG----TVVGRGAEVAGGARLDGAVIFDGVKIGAGAVIERSI
MAB_3611c|M.abscessus_ATCC_199 VAPGALVIGG----TVVGRGAEIGPGARLDGAVVFDGARIEAGAVVERSI
MSMEG_5078|M.smegmatis_MC2_155 LAPAKFVNGGSAQESVVGAGSIIS-AASVRNSVLSSNVVVDDGAIVEGSV
::*. .: ** :*** *: :. .. : .:*: ... : *.::* *:
MLBr_00753|M.leprae_Br4923 LGFGVRIGPRALIRDGVIGDGADIG-ARCELLRGARVWPGVSIPDGGIRY
MAV_4228|M.avium_104 VGFGARIGPRALIRDGVIGDGADIG-ARCELLRGARVWPGVSIPDGGIRY
MMAR_1277|M.marinum_M IGFGARIGPRALVRDGVIGDGADIG-ARCELLRGARVWPGVSIPDGGIRY
MUL_2610|M.ulcerans_Agy99 IGFGARIGPRALVRDGVIGDGADIG-ARCELLRGARVWPGVSIPDGGIRY
Mb3292c|M.bovis_AF2122/97 IGFGARIGPRALIRDGVIGDGADIG-ARCELLSGARVWPGVFLPDGGIRY
Rv3264c|M.tuberculosis_H37Rv IGFGARIGPRALIRDGVIGDGADIG-ARCELLSGARVWPGVFLPDGGIRY
TH_0779|M.thermoresistible__bu IGFGARIGPRALIRDGVIGDGADIG-ARCELLRGARVWPGVAIPDGGIRF
Mflv_4734|M.gilvum_PYR-GCK IGFGAHIGPRALIRDGVIGDGADIG-ARCELLRGLRVWPGITIPDGGIRF
Mvan_1729|M.vanbaalenii_PYR-1 IGFGARIGPRALIRDGVIGDGADIG-ARCELLRGARVWPGITIPDGGIRF
MAB_3611c|M.abscessus_ATCC_199 IGFGARIGPRALVRDGVIGDGADIG-ARCELLRGARVWPGITIPDGGIRY
MSMEG_5078|M.smegmatis_MC2_155 LMPGVRIGRGAVVRHAILDKNVVVGPGEMVGVDLDKDRERFAISAGGVVA
: *.:** *::*..::.... :* .. : : . :. **:
MLBr_00753|M.leprae_Br4923 SSDV---
MAV_4228|M.avium_104 SSDV---
MMAR_1277|M.marinum_M SSDV---
MUL_2610|M.ulcerans_Agy99 SSDV---
Mb3292c|M.bovis_AF2122/97 SSDV---
Rv3264c|M.tuberculosis_H37Rv SSDV---
TH_0779|M.thermoresistible__bu STDV---
Mflv_4734|M.gilvum_PYR-GCK STDI---
Mvan_1729|M.vanbaalenii_PYR-1 STDI---
MAB_3611c|M.abscessus_ATCC_199 SSDV---
MSMEG_5078|M.smegmatis_MC2_155 VGKGVWI
.