For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MRASPHVLGIVLAGGEGKRLYPLTADRAKPAVPFGGAYRLIDFVLSNLVNARFLRICVLTQYKSHSLDRH ISQNWRLSGLAGEYITPVPAQQRLGPRWYTGSADAIHQSLNLIFDEDPEYIVVFGADHVYRMDPEQMLDF HIESGAAVTVAGIRVPRTEASAFGCIDADDTGRIREFVEKPANPPGTPDDPDAAFVSMGNYIFTTKELID VIRADADDDHSDHDMGGDIIPRLVADGRAAVYDFNTNLVPGATERDHAYWRDVGTLDAFYDAHMDLVSVH PVFNLYNRRWPIRGESENLAPAKFVNGGSAQESVVGAGSIVSAASVRNSVLSSNVVIDDGAVVEGSVLMP GVRVGRGAVVRHAILDKNVVVGVGEQVGVDIERDRERFNVSAGGVVAVGKGVWI
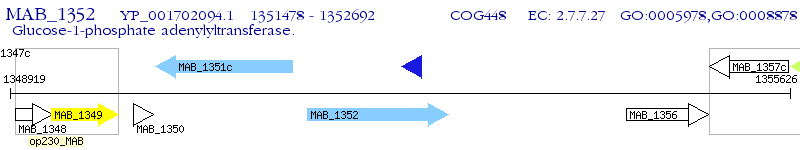
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_1352 | glgC | - | 100% (404) | glucose-1-phosphate adenylyltransferase |
| M. abscessus ATCC 19977 | MAB_3611c | - | 5e-23 | 27.61% (373) | putative sugar-phosphate nucleotidyl transferase |
| M. abscessus ATCC 19977 | MAB_4113 | - | 3e-05 | 23.21% (280) | glucose-1-phosphate thymidylyltransferase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1245 | glgC | 0.0 | 89.85% (404) | glucose-1-phosphate adenylyltransferase |
| M. gilvum PYR-GCK | Mflv_2193 | glgC | 0.0 | 88.61% (404) | glucose-1-phosphate adenylyltransferase |
| M. tuberculosis H37Rv | Rv1213 | glgC | 0.0 | 89.85% (404) | glucose-1-phosphate adenylyltransferase |
| M. leprae Br4923 | MLBr_01069 | glgC | 0.0 | 85.89% (404) | glucose-1-phosphate adenylyltransferase |
| M. marinum M | MMAR_4225 | glgC | 0.0 | 89.36% (404) | glucose-1-phosphate adenylyltransferase GlgC |
| M. avium 104 | MAV_1358 | glgC | 0.0 | 89.70% (398) | glucose-1-phosphate adenylyltransferase |
| M. smegmatis MC2 155 | MSMEG_5078 | glgC | 0.0 | 90.35% (404) | glucose-1-phosphate adenylyltransferase |
| M. thermoresistible (build 8) | TH_0549 | glgC | 0.0 | 89.70% (398) | glucose-1-phosphate adenylyltransferase |
| M. ulcerans Agy99 | MUL_4526 | glgC | 0.0 | 88.86% (404) | glucose-1-phosphate adenylyltransferase |
| M. vanbaalenii PYR-1 | Mvan_4503 | glgC | 0.0 | 88.86% (404) | glucose-1-phosphate adenylyltransferase |
CLUSTAL 2.0.9 multiple sequence alignment
MLBr_01069|M.leprae_Br4923 MREVPQVLGIVLAGGEGKRLYPLTADRAKPAVPFGGAYRLVDFVLSNLVN
MAV_1358|M.avium_104 ------MLGIVLAGGEGKRLYPLTADRAKPAVPFGGAYRLIDFVLSNLVN
Mb1245|M.bovis_AF2122/97 MREVPHVLGIVLAGGEGKRLYPLTADRAKPAVPFGGAYRLIDFVLSNLVN
Rv1213|M.tuberculosis_H37Rv MREVPHVLGIVLAGGEGKRLYPLTADRAKPAVPFGGAYRLIDFVLSNLVN
MMAR_4225|M.marinum_M MREAPHVLGIVLAGGEGKRLYPLTADRAKPAVPFGGAYRLIDFVLSNLVN
MUL_4526|M.ulcerans_Agy99 MREAPHVLGIVLAGGEGKRLYPLTADRAKPAVPFGGAYRLIDFVLSNLVN
Mflv_2193|M.gilvum_PYR-GCK MREAPHVLGIVLAGGEGKRLYPLTADRAKPAVPFGGGYRLIDFVLSNLVN
Mvan_4503|M.vanbaalenii_PYR-1 MREAPHVLGIVLAGGEGKRLYPLTADRAKPAVPFGGGYRLIDFVLSNLVN
MSMEG_5078|M.smegmatis_MC2_155 MRELPHVLGIVLAGGEGKRLYPLTADRAKPAVPFGGAYRLIDFVLSNLVN
TH_0549|M.thermoresistible__bu ------VLGIVLAGGEGKRLYPLTADRAKPAVPFGGAYRLIDFVLSNLVN
MAB_1352|M.abscessus_ATCC_1997 MRASPHVLGIVLAGGEGKRLYPLTADRAKPAVPFGGAYRLIDFVLSNLVN
:*****************************.***:*********
MLBr_01069|M.leprae_Br4923 ARYLRICVLTQYKSHSLDRHISQNWRLSGLAGEYITPVPAQQRFGPHWYT
MAV_1358|M.avium_104 ARYLRICVLTQYKSHSLDRHISQNWRLSGLAGEYITPVPAQQRLGPRWYT
Mb1245|M.bovis_AF2122/97 ARYLRICVLTQYKSHSLDRHISQNWRLSGLAGEYITPVPAQQRLGPRWYT
Rv1213|M.tuberculosis_H37Rv ARYLRICVLTQYKSHSLDRHISQNWRLSGLAGEYITPVPAQQRLGPRWYT
MMAR_4225|M.marinum_M ARYLRICVLTQYKSHSLDRHISQNWRLSGLAGEYITPVPAQQRLGPRWYT
MUL_4526|M.ulcerans_Agy99 ARYLRICVLTQYKSHSLDRHISQNWRLSGLAGEYITPVPAQQRLGPRWYT
Mflv_2193|M.gilvum_PYR-GCK ARFLRICVLTQYKSHSLDRHISQNWRLSGLAGEYITPVPAQQRLGPRWYT
Mvan_4503|M.vanbaalenii_PYR-1 ARFLRICVLTQYKSHSLDRHISQNWRLSGLAGEYITPVPAQQRLGPRWYT
MSMEG_5078|M.smegmatis_MC2_155 ARYLRICVLTQYKSHSLDRHISQNWRLSGLAGEYITPVPAQQRLGPRWYT
TH_0549|M.thermoresistible__bu ARFLRICVLTQYKSHSLDRHISQNWRLSGLAGEYITPVPAQQRLGPRWYT
MAB_1352|M.abscessus_ATCC_1997 ARFLRICVLTQYKSHSLDRHISQNWRLSGLAGEYITPVPAQQRLGPRWYT
**:****************************************:**:***
MLBr_01069|M.leprae_Br4923 GSADAIYQSLNLIYDEDPDYLVVFGADHVYRMDPEQMLRFHIGSGAGATV
MAV_1358|M.avium_104 GSADAIYQSLNLIYDEDPDYIVVFGADHVYRMDPEQMVRFHIDSGAGATV
Mb1245|M.bovis_AF2122/97 GSADAIYQSLNLIYDEDPDYIVVFGADHVYRMDPEQMVRFHIDSGAGATV
Rv1213|M.tuberculosis_H37Rv GSADAIYQSLNLIYDEDPDYIVVFGADHVYRMDPEQMVRFHIDSGAGATV
MMAR_4225|M.marinum_M GSADAIYQSLNLIYDEDPDYIVVFGADHVYRMDPEQMVRFHIDSGAGVTV
MUL_4526|M.ulcerans_Agy99 GSADAIYQSLNLIYDEDPDYIVVFGADHVYRMDPEQMVRFHIDSGAGVTV
Mflv_2193|M.gilvum_PYR-GCK GSADAIYQSMNLIYDEDPDYIVIFGADHVYRMDPEQMVQQHIESGAGATV
Mvan_4503|M.vanbaalenii_PYR-1 GSADAIYQSMNLIYDEDPDYIVIFGADHVYRMDPEQMVQQHIESGAGATV
MSMEG_5078|M.smegmatis_MC2_155 GSADAIYQSLNLIYDEDPDYIIIFGADHVYRMDPEQMMQFHIESGAGATV
TH_0549|M.thermoresistible__bu GSADAIYQSMNLIYDEDPDYIVVFGADHVYRMDPEQMLRFHIESGAGATV
MAB_1352|M.abscessus_ATCC_1997 GSADAIHQSLNLIFDEDPEYIVVFGADHVYRMDPEQMLDFHIESGAAVTV
******:**:***:****:*:::**************: ** ***..**
MLBr_01069|M.leprae_Br4923 AGIRVPRSDATAFGCIDADDSGRIRRFTEKPLKPPGTPDDPDKTFVSMGN
MAV_1358|M.avium_104 AGIRVPRSEATAFGCIDSDESGRIRKFVEKPLDPPGTPDDPETTFVSMGN
Mb1245|M.bovis_AF2122/97 AGIRVPRENATAFGCIDADDSGRIRSFVEKPLEPPGTPDDPDTTFVSMGN
Rv1213|M.tuberculosis_H37Rv AGIRVPRENATAFGCIDADDSGRIRSFVEKPLEPPGTPDDPDTTFVSMGN
MMAR_4225|M.marinum_M AGIRVPRADASAFGCIDADDAGTIRDFVEKPLEPPGTPDDPTSTFVSMGN
MUL_4526|M.ulcerans_Agy99 AGIRVPRADASAFGCIDADDAGTIRDFVEKPLEPPGTPDDPTSTFVSMGN
Mflv_2193|M.gilvum_PYR-GCK AGIRVPRSEASAFGCIDADDSGRIRGWVEKPADPPGTPDDPEMTFASMGN
Mvan_4503|M.vanbaalenii_PYR-1 AGIRVPRAEASAFGCIDSDDSGRIRGFIEKPADPPGTPDDPEQTFVSMGN
MSMEG_5078|M.smegmatis_MC2_155 AGIRVPRSEASAFGCIDADESGRIREFIEKPADPPGTPDDPEQTFVSMGN
TH_0549|M.thermoresistible__bu AGIRVPRKEATAFGVIDADESGRIRAFVEKPAEPPAVPDNPDEAFASMGN
MAB_1352|M.abscessus_ATCC_1997 AGIRVPRTEASAFGCIDADDTGRIREFVEKPANPPGTPDDPDAAFVSMGN
******* :*:*** **:*::* ** : *** .**..**:* :*.****
MLBr_01069|M.leprae_Br4923 YIFTTKVLVDAIRADADDDHSYHDMGGDILPRLVDGGMAAVYDFSQNEVP
MAV_1358|M.avium_104 YIFTTKVLIDAIRADADDDHSDHDMGGDIIPRLVDDGMAAVYDFSDNEVP
Mb1245|M.bovis_AF2122/97 YIFTTKVLIDAIRADADDDHSDHDMGGDIVPRLVADGMAAVYDFSDNEVP
Rv1213|M.tuberculosis_H37Rv YIFTTKVLIDAIRADADDDHSDHDMGGDIVPRLVADGMAAVYDFSDNEVP
MMAR_4225|M.marinum_M YVFTTKVLIDAIRADADDDHSDHDMGGDIIPRLVADGMAAVYDFSNNEVP
MUL_4526|M.ulcerans_Agy99 YVFTTKVLIDAIRADADDDHSDHDMGGDIIPRLVADGMAVVYDFSNNEVP
Mflv_2193|M.gilvum_PYR-GCK YIFTTKVLIDAIRADAEDDDSDHDMGGDIIPRLVADGMASVYDFNNNEVP
Mvan_4503|M.vanbaalenii_PYR-1 YIFTTKVLIDAIRADAEDDDSDHDMGGDIIPRLVADGMAAVYDFNDNEVP
MSMEG_5078|M.smegmatis_MC2_155 YIFTTKVLIDAIRADADDDHSDHDMGGDIIPRLVADGMAAVYDFKNNEVP
TH_0549|M.thermoresistible__bu YIFTTKVLIDAIKADAEDDHSDHDMGGDIIPRLVEDGMAAVYDFNDNEVP
MAB_1352|M.abscessus_ATCC_1997 YIFTTKELIDVIRADADDDHSDHDMGGDIIPRLVADGRAAVYDFNTNLVP
*:**** *:*.*:***:**.* *******:**** .* * ****. * **
MLBr_01069|M.leprae_Br4923 GATDWDRAYWRDVGTLDAFYDAHMDLVSLRPVFNLYNKRWPIRGESENLA
MAV_1358|M.avium_104 GATDRDRGYWRDVGTLDAFYDAHMDLVSVHPVFNLYNKRWPIRGESENLA
Mb1245|M.bovis_AF2122/97 GATDRDRAYWRDVGTLDAFYDAHMDLVSVHPVFNLYNKRWPIRGESENLA
Rv1213|M.tuberculosis_H37Rv GATDRDRAYWRDVGTLDAFYDAHMDLVSVHPVFNLYNKRWPIRGESENLA
MMAR_4225|M.marinum_M GATDRDRAYWRDVGTLDAFYDAHMDLVSVHPVFNLYNKRWPIRGESENLA
MUL_4526|M.ulcerans_Agy99 GATDRDRAYWRDVGTLDAFYDAHMDLVSVHPVFNLYNKRWPIRGESENLA
Mflv_2193|M.gilvum_PYR-GCK GATERDHGYWRDVGTLDAFYDAHMDLVSVHPIFNLYNKRWPIRGGAENLA
Mvan_4503|M.vanbaalenii_PYR-1 GATERDHGYWRDVGTLDAFYDAHMDLVSVHPVFNLYNKRWPIRGGAENLA
MSMEG_5078|M.smegmatis_MC2_155 GATERDHGYWRDVGTLDAFYDAHMDLVSVHPVFNLYNKRWPIRGESENLA
TH_0549|M.thermoresistible__bu GATERDHGYWRDVGTLDAFYDAHMDLVSVHPVFNLYNRRWPIRGATENLA
MAB_1352|M.abscessus_ATCC_1997 GATERDHAYWRDVGTLDAFYDAHMDLVSVHPVFNLYNRRWPIRGESENLA
***: *:.********************::*:*****:****** :****
MLBr_01069|M.leprae_Br4923 PAKFVNGGSVQESVVGAGSIISAASVRNSVLSSNVVVDNGAIVEGSVIMP
MAV_1358|M.avium_104 PAKFVNGGSAQESVVGAGSIISAASVRNSVLSSNVVVDDGAIVEGSVIMP
Mb1245|M.bovis_AF2122/97 PAKFVNGGSAQESVVGAGSIISAASVRNSVLSSNVVVDDGAIVEGSVIMP
Rv1213|M.tuberculosis_H37Rv PAKFVNGGSAQESVVGAGSIISAASVRNSVLSSNVVVDDGAIVEGSVIMP
MMAR_4225|M.marinum_M PAKFVNGGSAQESVVGAGSIISAASVRNSVLSSNVVVEDGAIVEGSVIMP
MUL_4526|M.ulcerans_Agy99 PAKFVNGGSAQESVVGAGSIISAASVRNSVLSSNVVVEDGAIVEGSVIMP
Mflv_2193|M.gilvum_PYR-GCK PAKFVNGGSAQESVVGAGSIISAASVRNSVLSSNVAIDDGAIVEGSVIMP
Mvan_4503|M.vanbaalenii_PYR-1 PAKFVNGGSAQESVVGAGSIISAASVRNSVLSSNVVIDDGAIVEGSVIMP
MSMEG_5078|M.smegmatis_MC2_155 PAKFVNGGSAQESVVGAGSIISAASVRNSVLSSNVVVDDGAIVEGSVLMP
TH_0549|M.thermoresistible__bu PAKFVNGGSAQESVVGAGSIISAASVRNSVLSSNVVIDDGAIVEGSVLMP
MAB_1352|M.abscessus_ATCC_1997 PAKFVNGGSAQESVVGAGSIVSAASVRNSVLSSNVVIDDGAVVEGSVLMP
*********.**********:**************.:::**:*****:**
MLBr_01069|M.leprae_Br4923 GARVGRGAVIRHAILDKNVVVGPGEMVGVDPERDREHFAISAGGVVVVGK
MAV_1358|M.avium_104 GARVGRGAVVRHAILDKNVVVGPGEMVGVDLERDRERFAISAGGVVAVGK
Mb1245|M.bovis_AF2122/97 GTRVGRGAVVRHAILDKNVVVGPGEMVGVDLEKDRERFAISAGGVVAVGK
Rv1213|M.tuberculosis_H37Rv GTRVGRGAVVRHAILDKNVVVGPGEMVGVDLEKDRERFAISAGGVVAVGK
MMAR_4225|M.marinum_M GARVGRGAVVRHAILDKNVVVGPGEMVGVDLDKDRERFAISAGGVVAVGK
MUL_4526|M.ulcerans_Agy99 GARVGRDAVVRHAILDKNVVVGPGEMVGVDLDKDRERFAISAGGVVAVGK
Mflv_2193|M.gilvum_PYR-GCK GVRIGRGAVVRHAILDKNVVVGPGEMVGVDLDKDRERFTVSAGGVVAVGK
Mvan_4503|M.vanbaalenii_PYR-1 GARIGRGAVVRHAILDKNVVVGPGEMVGVDLDKDRERFSISAGGVVAVGK
MSMEG_5078|M.smegmatis_MC2_155 GVRIGRGAVVRHAILDKNVVVGPGEMVGVDLDKDRERFAISAGGVVAVGK
TH_0549|M.thermoresistible__bu GVRVGRGAVVRRSILDKNVVVGPGEMVGVDLEKDRERFAISAGGVVTVGK
MAB_1352|M.abscessus_ATCC_1997 GVRVGRGAVVRHAILDKNVVVGVGEQVGVDIERDRERFNVSAGGVVAVGK
*.*:**.**:*::********* ** **** ::***:* :******.***
MLBr_01069|M.leprae_Br4923 GVWI
MAV_1358|M.avium_104 GVWI
Mb1245|M.bovis_AF2122/97 GVWI
Rv1213|M.tuberculosis_H37Rv GVWI
MMAR_4225|M.marinum_M GVWI
MUL_4526|M.ulcerans_Agy99 GVWI
Mflv_2193|M.gilvum_PYR-GCK GVWI
Mvan_4503|M.vanbaalenii_PYR-1 GVWI
MSMEG_5078|M.smegmatis_MC2_155 GVWI
TH_0549|M.thermoresistible__bu GVWI
MAB_1352|M.abscessus_ATCC_1997 GVWI
****