For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TFRLRNIPLLSRVGLDRADELRSNPEELAKGWAEAGLITLDVRGRVNIVDGQVVIEDAARIGDQPPEHAV FLGRIPGGRHVWAVRADLDEDSAPLLDLRRSGQLFDDTSAALLATAMAMLAWHDNAGYSPVDGSPTIPAK GGWVRVNSATGQEEFPRTDPAIICLVHDGGDRAVLGRQKFWPERMFSLLAGFVEAGESLEACVAREVAEE VGLTVTDVQYLGSQPWPFPRSIMLGFHAIGDPSQPFAFNDGEIAEADWFTRAEVRSALEAGDWTTASDSR LMLPGSISIAREIVESWAYAGEEPPAGRP*
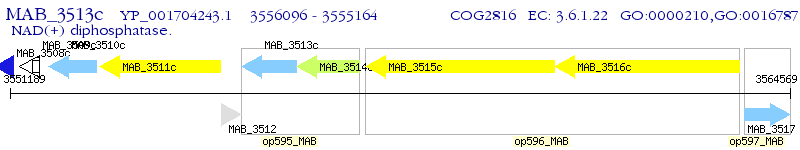
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_3513c | nudC | - | 100% (310) | NADH pyrophosphatase |
| M. abscessus ATCC 19977 | MAB_4342c | - | 5e-06 | 32.41% (108) | putative MutT/nudix family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3224c | nudC | 1e-115 | 64.57% (302) | NADH pyrophosphatase |
| M. gilvum PYR-GCK | Mflv_4662 | nudC | 1e-123 | 67.88% (302) | NADH pyrophosphatase |
| M. tuberculosis H37Rv | Rv3199c | nudC | 1e-114 | 64.24% (302) | NADH pyrophosphatase |
| M. leprae Br4923 | - | - | - | - | - |
| M. marinum M | MMAR_1361 | nudC | 1e-119 | 66.12% (304) | NADH pyrophosphatase NudC |
| M. avium 104 | MAV_4145 | nudC | 1e-117 | 65.23% (302) | NADH pyrophosphatase |
| M. smegmatis MC2 155 | MSMEG_1946 | nudC | 1e-124 | 69.31% (303) | NADH pyrophosphatase |
| M. thermoresistible (build 8) | TH_1137 | - | 5e-94 | 71.62% (222) | PROBABLE NADH PYROPHOSPHATASE NUDC (NAD+ DIPHOSPHATASE) (NAD+ |
| M. ulcerans Agy99 | MUL_2518 | nudC | 1e-119 | 66.12% (304) | NADH pyrophosphatase |
| M. vanbaalenii PYR-1 | Mvan_1805 | nudC | 1e-118 | 66.45% (301) | NADH pyrophosphatase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_4662|M.gilvum_PYR-GCK -----MTPFRLRNIPLLSRVGADRADTLRTDIDAAVAGWSEALLLRVDRR
Mvan_1805|M.vanbaalenii_PYR-1 -----MTAFRLRNVPLLSRVGADRADTLRTDIDAAVAGWPDALLLRVDRR
MSMEG_1946|M.smegmatis_MC2_155 --MSEHRTFGLRNVPLLSRVGADRADTLRTDVDAALAGWPDALVLRVDRR
TH_1137|M.thermoresistible__bu --------------------------------------------------
Mb3224c|M.bovis_AF2122/97 MTNVSGVDFQLRSVPLLSRVGADRADRLRTDMEAAAAGWPGAALLRVDSR
Rv3199c|M.tuberculosis_H37Rv MTNVSGVDFQLRSVPLLSRVGADRADRLRTDMEAAAAGWPGAALLRVDSR
MAV_4145|M.avium_104 ---MAIADFQLRSVPLLSRVGADRADQLRTDVEAAAAGWADAALLRVDSR
MMAR_1361|M.marinum_M ------MDFQLREVPLLSRIGADRADHLRTDVEAAAAGWADAALMRVDSR
MUL_2518|M.ulcerans_Agy99 --MRASVDFQLREVPLLSRIGADRADHLRTDVEAAAAGWADAALMRVDSR
MAB_3513c|M.abscessus_ATCC_199 ------MTFRLRNIPLLSRVGLDRADELRSNPEELAKGWAEAGLITLDVR
Mflv_4662|M.gilvum_PYR-GCK NQVLISDGRAVLNRAVSVGPSPPENAVFLGKLDDGRHVWAVRSDLQPPED
Mvan_1805|M.vanbaalenii_PYR-1 NQVLISGGRAVLNSASSAGAAPPEDAVFLGRLDDGRHVWAVRSELEPPED
MSMEG_1946|M.smegmatis_MC2_155 NQVLIANGQVVLGEAGALGDRPPEHAVFLGRLQDGRHVWGIRADLEAPED
TH_1137|M.thermoresistible__bu -------------------------------------VWAVRAALEPPAD
Mb3224c|M.bovis_AF2122/97 NRVLVANGRVLLGAAIELADKPPPEAVFLGRVEGGRHVWAVRAALQPIAD
Rv3199c|M.tuberculosis_H37Rv NRVLVANGRVLLGAAIELADKPPPEAVFLGRVEGGRHVWAVRAALQPIAD
MAV_4145|M.avium_104 NQALVADGRVVLGAAAELGDKPPPEAVFLGRLEDGRHVWAIRGALQAPDD
MMAR_1361|M.marinum_M NRVLVADGRVVLSPAADFAEKPPPEAVFLGCLEGGRHVWAVRAALEAPED
MUL_2518|M.ulcerans_Agy99 NRVLVADGRVVLSPAADFAEKPPPEAVFLGCLEGGRHVWAVRAALEAPED
MAB_3513c|M.abscessus_ATCC_199 GRVNIVDGQVVIEDAARIGDQPPEHAVFLGRIPGGRHVWAVRADLD--ED
**.:*. *: *
Mflv_4662|M.gilvum_PYR-GCK ETAPVQVLDLRRTGDVFDDVSAQLVATATALLNWHDSARFSPVDGAPTTP
Mvan_1805|M.vanbaalenii_PYR-1 PDAATEVLDLRRTGDVFDDVSAQLVATATALLNWHDHARFSPVDGAPTRP
MSMEG_1946|M.smegmatis_MC2_155 ADLGTEVLDLRRAGQIFDDTSAQLVATATALLNWHDNARHSAIDGAPTRP
TH_1137|M.thermoresistible__bu R-PDATVLDLRRTGHIFDDVSAQLVATATALLNWHDNAKFSAIDGAPTKP
Mb3224c|M.bovis_AF2122/97 PDIPAEAVDLRGLGRIMDDTSSQLVSSASALLNWHDNARFSALDGAPTKP
Rv3199c|M.tuberculosis_H37Rv PDIPAEAVDLRGLGRIMDDTSSQLVSSASALLNWHDNARFSALDGAPTKP
MAV_4145|M.avium_104 PKVRAEVVNLRSLGPIFDDTSSQLMSSAVALLNWHERSRFSSVDGSPTRP
MMAR_1361|M.marinum_M PGAQSEVADLRKLGSIIDDTSSQLVASAMALLNWHDGSRFSPVDGSPTKP
MUL_2518|M.ulcerans_Agy99 PGAQSEVADLRKLGSIIDDTSSQLVASAMALLNWHDGSRFSPVDGSPTKP
MAB_3513c|M.abscessus_ATCC_199 S---APLLDLRRSGQLFDDTSAALLATAMAMLAWHDNAGYSPVDGSPTIP
:** * ::**.*: *:::* *:* **: : .*.:**:** *
Mflv_4662|M.gilvum_PYR-GCK IKSGWARINPVTGQEEFPRIDPAVICLVHDGHDRAVLARQTVWPPRLFSI
Mvan_1805|M.vanbaalenii_PYR-1 VKSGWARINPVTGHEEFPRIDPAVICLVHDGHDRAVLARQTAWPPRLFSI
MSMEG_1946|M.smegmatis_MC2_155 AKGGWSRVNPLTGHEEFPRIDPAIICLVHDGHDRAVLARQTLWPERLFSI
TH_1137|M.thermoresistible__bu VNGGWSRVNLVNGHEEFPRIDPAVICLVHDGHDRCVLARQRLWPERFFSL
Mb3224c|M.bovis_AF2122/97 ARAGWSRVNPITGHEEFPRIDPAVICLVHDGADRAVLARQAAWPERMFSL
Rv3199c|M.tuberculosis_H37Rv ARAGWSRVNPITGHEEFPRIDPAVICLVHDGADRAVLARQAAWPERMFSL
MAV_4145|M.avium_104 ARAGWSRVNPVTGHEEFPRIDPAVICLVHDGGDRAVLARQAVWPERMFSL
MMAR_1361|M.marinum_M ARAGWSRVNSLTGHEEFPRIDPAVICLVHDGGDRVVLARQAIWPHRMFSL
MUL_2518|M.ulcerans_Agy99 ARAGWSRVNSLTGHEEFPRIDPAVICLVHDGGDRVVLARQAIWPHRMFSL
MAB_3513c|M.abscessus_ATCC_199 AKGGWVRVNSATGQEEFPRTDPAIICLVHDGGDRAVLGRQKFWPERMFSL
..** *:* .*:***** ***:******* ** **.** ** *:**:
Mflv_4662|M.gilvum_PYR-GCK LAGFVEAGESFESCVVREIAEEIGLTVTDVEYLGSQPWPFPRSLMVGFHA
Mvan_1805|M.vanbaalenii_PYR-1 LAGFVEAGESFESCVVREIAEEIGLTVTDVEYLGSQPWPFPRSLMVGFHA
MSMEG_1946|M.smegmatis_MC2_155 LAGFVEAGESFETCVQREIAEEIGLTVTDVQYLGSQPWPFPRSLMVGFHA
TH_1137|M.thermoresistible__bu VAGFVEAGESFESCVVREIAEELGLTVTDVEYLGSQPWPFPRSLMVGFHA
Mb3224c|M.bovis_AF2122/97 LAGFVEAGESFEVCVAREIREEIGLTVRDVRYLGSQPWPFPRSLMVGFHA
Rv3199c|M.tuberculosis_H37Rv LAGFVEAGESFEVCVAREIREEIGLTVRDVRYLGSQQWPFPRSLMVGFHA
MAV_4145|M.avium_104 LAGFVEAGESFEVCVAREVREEIGLTVRDVRYLGSQPWPFPRSLMVGFHA
MMAR_1361|M.marinum_M LAGFVEAGESFEVCVAREIREEIGLTVRDVRYLGSQPWPLPRSLMVGFHA
MUL_2518|M.ulcerans_Agy99 LAGFVEAGESFEVCVAREIREEIGLTVRDVRYLGSQPWPLPRSLMVGFHA
MAB_3513c|M.abscessus_ATCC_199 LAGFVEAGESLEACVAREVAEEVGLTVTDVQYLGSQPWPFPRSIMLGFHA
:*********:* ** **: **:**** **.***** **:***:*:****
Mflv_4662|M.gilvum_PYR-GCK VGDPEQPFSFNDGEIAEAQWFTRAEIRDALDQGDWSSDSSSRLLLPGSIS
Mvan_1805|M.vanbaalenii_PYR-1 IGDPEQPFAFNDGEIAEAAWFTRAEIREALAQGDWGSGGTSRLLLPGSIS
MSMEG_1946|M.smegmatis_MC2_155 IGDPEQPFSYNDGEIAEAAWFTRDEIRAALDQGDWNSDSPSRLLLPGSIS
TH_1137|M.thermoresistible__bu VGDPDQPFSFNDGEIVEAAWFTRDEVREALAAGDWSSDSSSRLLLPGSIS
Mb3224c|M.bovis_AF2122/97 LGDPDEEFSFSDGEIAEAAWFTRDEVRAALAAGDWSSASESKLLLPGSIS
Rv3199c|M.tuberculosis_H37Rv LGDPDEEFSFSDGEIAEAAWFTRDEVRAALAAGDWSSASESKLLLPGSIS
MAV_4145|M.avium_104 VADPAQDFAFNDGEIAEAAWFTRDEVRAALAAGDWSSDSESKLLLPGSIS
MMAR_1361|M.marinum_M VGDPDEDFSFNDGEIAEAAWFTRDEVRAALAVGDWTSASDSKLLLPGSIS
MUL_2518|M.ulcerans_Agy99 VGDPDEDFSFNDGEIAEAAWFTRDEVRAALAVGDWTSASDSKLLLPGSIS
MAB_3513c|M.abscessus_ATCC_199 IGDPSQPFAFNDGEIAEADWFTRAEVRSALEAGDWTTASDSRLMLPGSIS
:.** : *::.****.** **** *:* ** *** : . *:*:******
Mflv_4662|M.gilvum_PYR-GCK IAREIIESWAALD--------
Mvan_1805|M.vanbaalenii_PYR-1 IAREIIESWVERD--------
MSMEG_1946|M.smegmatis_MC2_155 IAREIIESWAALD--------
TH_1137|M.thermoresistible__bu IAREIIESWAELD--------
Mb3224c|M.bovis_AF2122/97 IARVIIESWAACE--------
Rv3199c|M.tuberculosis_H37Rv IARVIIESWAACE--------
MAV_4145|M.avium_104 IARVIIESWAALD--------
MMAR_1361|M.marinum_M IARVIIESWAAARD-------
MUL_2518|M.ulcerans_Agy99 IARVIIESWAAARD-------
MAB_3513c|M.abscessus_ATCC_199 IAREIVESWAYAGEEPPAGRP
*** *:***.