For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
ISLDKVTKLYKSSARPALDNVSLEIDKGEFVFLIGPSGSGKSTFMRLLLAEELPTKGDVQVSKFHVNKLR GREIPHLRQTIGCVFQDFRLLQQKTVYENVAFALEVIGKQTDIINRVVPEVLEMVNLTGKASRLPSELSG GEQQRVGIARAFVNRPLVLLADEPTGNLDPETSEDIMALLERINRTGTTVLMATHDHHIVDSMRQRVIEL ELGKLIRDEQRGVYGVDR*
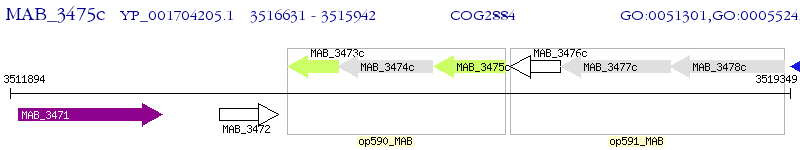
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_3475c | - | - | 100% (229) | putative cell division ATP-binding protein FtsE |
| M. abscessus ATCC 19977 | MAB_2803c | - | 5e-42 | 43.18% (220) | methionine import ATP-binding protein metN |
| M. abscessus ATCC 19977 | MAB_1655 | - | 1e-32 | 37.50% (200) | sulfate ABC transporter, ATP-binding protein SysA |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3129c | ftsE | 1e-110 | 85.59% (229) | putative cell division ATP-binding protein FTSE (septation |
| M. gilvum PYR-GCK | Mflv_4448 | - | 1e-110 | 85.59% (229) | type II secretory pathway family protein |
| M. tuberculosis H37Rv | Rv3102c | ftsE | 1e-110 | 85.59% (229) | putative cell division ATP-binding protein FTSE (septation |
| M. leprae Br4923 | MLBr_00669 | ftsE | 1e-106 | 82.10% (229) | putative cell division ATP-binding protein |
| M. marinum M | MMAR_1530 | ftsE | 1e-109 | 84.28% (229) | cell division ATP-binding protein FtsE |
| M. avium 104 | MAV_4003 | ftsE | 1e-108 | 84.28% (229) | cell division ATP-binding protein FtsE |
| M. smegmatis MC2 155 | MSMEG_2089 | ftsE | 1e-110 | 86.03% (229) | cell division ATP-binding protein FtsE |
| M. thermoresistible (build 8) | TH_3185 | - | 1e-110 | 84.72% (229) | PUTATIVE cell division ATP-binding protein FtsE |
| M. ulcerans Agy99 | MUL_2409 | ftsE | 1e-109 | 84.28% (229) | cell division ATP-binding protein FtsE |
| M. vanbaalenii PYR-1 | Mvan_1914 | - | 1e-110 | 86.03% (229) | type II secretory pathway family protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3129c|M.bovis_AF2122/97 -MITLDHVTKQYKSSARPALDDINVKIDKGEFVFLIGPSGSGKSTFMRLL
Rv3102c|M.tuberculosis_H37Rv -MITLDHVTKQYKSSARPALDDINVKIDKGEFVFLIGPSGSGKSTFMRLL
MMAR_1530|M.marinum_M -MITLDHVSKQYKSSARPALDDINVKIDKGEFVFLIGPSGSGKSTFMRLL
MUL_2409|M.ulcerans_Agy99 -MITLDHVSKQYKSSARPALDDINVKIDKGEFVFLIGPSGSGKSTFMRLL
MLBr_00669|M.leprae_Br4923 -MITLDHVSKKYKSLARPALDNVNVKIDKGEFVFLIGPSGSGKSTFMRLL
MAV_4003|M.avium_104 MMITLDHVTKKYKASARPALEDVNVKIDKGEFVFLIGPSGSGKSTFMRLL
TH_3185|M.thermoresistible__bu -VITLDRVSKQYKSSARPALDNVSVKIEKGEFVFLIGPSGSGKSTFMRLL
Mflv_4448|M.gilvum_PYR-GCK -MITLDRVSKQYKSSARPALDNVSVKIDKGEFVFLIGPSGSGKSTFMRLL
Mvan_1914|M.vanbaalenii_PYR-1 -MITLDRVSKQYKSSARPALDNVSLKIEKGEFVFLIGPSGSGKSTFMRLL
MSMEG_2089|M.smegmatis_MC2_155 -MITLDHVTKQYKSSARPALDDVSLKIDKGEFVFLIGPSGSGKSTFMRLL
MAB_3475c|M.abscessus_ATCC_199 -MISLDKVTKLYKSSARPALDNVSLEIDKGEFVFLIGPSGSGKSTFMRLL
:*:**:*:* **: *****:::.::*:**********************
Mb3129c|M.bovis_AF2122/97 LAAETPTSGDVRVSKFHVNKLRGRHVPKLRQVIGCVFQDFRLLQQKTVYD
Rv3102c|M.tuberculosis_H37Rv LAAETPTSGDVRVSKFHVNKLRGRHVPKLRQVIGCVFQDFRLLQQKTVYD
MMAR_1530|M.marinum_M LAAETPTSGDVRVSKFHVNKLRGRNVPKLRQVIGCVFQDFRLLQQKTVYE
MUL_2409|M.ulcerans_Agy99 LAAETPTSGDVRVSKFHVNKLRGRNVPKLRQVIGCVFQDFRLLQQKTVYE
MLBr_00669|M.leprae_Br4923 LGAETPTSGDVRVSKFHVNKLPGRHIPRLRQVIGCVFQDFRLLQQKTVYE
MAV_4003|M.avium_104 LGEENPTTGDIRVSKFHVNKLRGRHIPKLRQVIGTVFQDFRLLQQKTVYE
TH_3185|M.thermoresistible__bu LGEEQPTSGDIRVSKFHVNKLRGRHIPSLRQVIGCVFQDFRLLQQKTVFE
Mflv_4448|M.gilvum_PYR-GCK LAEENPSSGDIRVSKFHVNKLSGRHIPGLRQVIGCVFQDFRLLQQKTVFE
Mvan_1914|M.vanbaalenii_PYR-1 LAEEHPSSGDIRVSKFHVNKLSGRHIPALRQVIGCVFQDFRLLQQKTVFE
MSMEG_2089|M.smegmatis_MC2_155 LGADTPTSGDVRVSKFHVNKLPGRHIPSLRQVIGCVFQDFRLLQQKTVFE
MAB_3475c|M.abscessus_ATCC_199 LAEELPTKGDVQVSKFHVNKLRGREIPHLRQTIGCVFQDFRLLQQKTVYE
*. : *:.**::********* **.:* ***.** *************::
Mb3129c|M.bovis_AF2122/97 NVAFALEVIGKRTDAINRVVPEVLETVGLSGKANRLPDELSGGEQQRVAI
Rv3102c|M.tuberculosis_H37Rv NVAFALEVIGKRTDAINRVVPEVLETVGLSGKANRLPDELSGGEQQRVAI
MMAR_1530|M.marinum_M NVAFALEVIGKRADAINRVVPDVLETVGLSGKANRLPHELSGGEQQRVAI
MUL_2409|M.ulcerans_Agy99 NVAFALEVIGKRADAINRVVPDVLETVGLSGKANRLPHELSGGEQQRVAI
MLBr_00669|M.leprae_Br4923 NVAFALEVIGRRSDAINQVVPDVLETVGLSGKANRLPDELSGGEQQRVAI
MAV_4003|M.avium_104 NVAFALEVIGKRTDAINRVVPEVLETVGLSGKANRLPHELSGGEQQRVAI
TH_3185|M.thermoresistible__bu NVAFALEVIGKRREVIDRVVPDVLEMVGLSGKANRLPAELSGGEQQRVAI
Mflv_4448|M.gilvum_PYR-GCK NVAFALEVIGKRADTINRVVPDVLEMVGLSGKANRLPGELSGGEQQRVAI
Mvan_1914|M.vanbaalenii_PYR-1 NVAFALEVIGKRADTINRVVPDVLEMVGLSGKANRLPAELSGGEQQRVAI
MSMEG_2089|M.smegmatis_MC2_155 NVAFALEVIGKRADTINRVVPDVLEMVGLSGKANRLPSELSGGEQQRVAI
MAB_3475c|M.abscessus_ATCC_199 NVAFALEVIGKQTDIINRVVPEVLEMVNLTGKASRLPSELSGGEQQRVGI
**********:: : *::***:*** *.*:***.*** **********.*
Mb3129c|M.bovis_AF2122/97 ARAFVNRPLVLLADEPTGNLDPETSRDIMDLLERINRTGTTVLMATHDHH
Rv3102c|M.tuberculosis_H37Rv ARAFVNRPLVLLADEPTGNLDPETSRDIMDLLERINRTGTTVLMATHDHH
MMAR_1530|M.marinum_M ARAFVNRPLVLLADEPTGNLDPDTSKDIMDLLERINRTGTTVLMATHDHH
MUL_2409|M.ulcerans_Agy99 ARAFVNRPLVLLADEPTGNLDPDTSKDIMDLLERINRTGTTVLMATHDHH
MLBr_00669|M.leprae_Br4923 ARAFVNRPLVLLADEPTGNLDPDTSKDIMDLLERINRTGTTVLMATHDHH
MAV_4003|M.avium_104 ARAFVNRPLVLLADEPTGNLDPDTSKDIMDLLERINRTGTTVLMATHDHH
TH_3185|M.thermoresistible__bu ARAFVNRPLVLLADEPTGNLDPETSKDIMDLLERINRTGTTVLMATHDHH
Mflv_4448|M.gilvum_PYR-GCK ARAFVNRPLVLLADEPTGNLDPDTSKDIMDLLERINRTGTTVVMATHDHH
Mvan_1914|M.vanbaalenii_PYR-1 ARAFVNRPLVLLADEPTGNLDPETSKDIMDLLERINRTGTTVVMATHDHH
MSMEG_2089|M.smegmatis_MC2_155 ARAFVNRPLVLIADEPTGNLDPETSKDIMDLLERINRTGTTVVMATHDHH
MAB_3475c|M.abscessus_ATCC_199 ARAFVNRPLVLLADEPTGNLDPETSEDIMALLERINRTGTTVLMATHDHH
***********:**********:**.*** ************:*******
Mb3129c|M.bovis_AF2122/97 IVDSMRQRVVELSLGRLVRDEQRGVYGMDR
Rv3102c|M.tuberculosis_H37Rv IVDSMRQRVVELSLGRLVRDEQRGVYGMDR
MMAR_1530|M.marinum_M IVDSMRQRVVELSLGRLVRDEQRGVYGMDR
MUL_2409|M.ulcerans_Agy99 IVDSMRQRVVELSLGRLVRDEQRGVYGMDR
MLBr_00669|M.leprae_Br4923 IVDSMRQRVVELSLGRLVRDEWCGIYGMDR
MAV_4003|M.avium_104 IVDSMRQRVVELSLGRLVRDEQRGIYGMDR
TH_3185|M.thermoresistible__bu IVDSMRQRVVELSLGRLVRDEQRGIYGMDR
Mflv_4448|M.gilvum_PYR-GCK IVDSMRQRVVELELGRLIRDEQRGVYGMDR
Mvan_1914|M.vanbaalenii_PYR-1 IVDSMRQRVVELELGRLIRDEQRGVYGMDR
MSMEG_2089|M.smegmatis_MC2_155 IVDSMRQRVVELELGRLIRDEQRGVYGMDR
MAB_3475c|M.abscessus_ATCC_199 IVDSMRQRVIELELGKLIRDEQRGVYGVDR
*********:**.**:*:*** *:**:**