For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MPQKVVIAGGHGQIAAHLIRLLAARGDTAVALIRNPDHAADVEAWGGLPLVLDLESTDADDVAGALAGAD AAVFAAGAGPGSGPARKDTVDRAAAVLLATAAEQAQVHRFIQISAFGAGEPAPTEGDESWIAYVIAKTAA EEDLRTRDRLDWTILRPGLLTDTAPTGSVTLSASKIERGSVPRADVAAVVAELLGAPNTVHEILFLTEGT APIASAVAGL
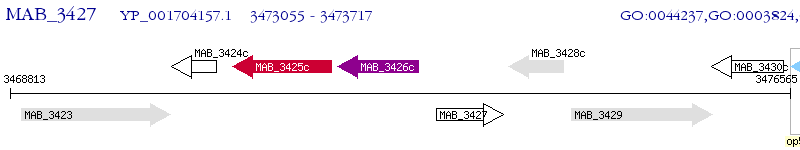
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_3427 | - | - | 100% (220) | hypothetical protein MAB_3427 |
| M. abscessus ATCC 19977 | MAB_1944c | - | 3e-05 | 29.75% (121) | hypothetical protein MAB_1944c |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_1761 | - | 1e-52 | 53.21% (218) | NAD-dependent epimerase/dehydratase |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. marinum M | MMAR_4327 | - | 3e-42 | 43.26% (215) | hypothetical protein MMAR_4327 |
| M. avium 104 | MAV_1263 | - | 1e-45 | 47.20% (214) | NAD dependent epimerase/dehydratase family protein |
| M. smegmatis MC2 155 | MSMEG_2941 | - | 6e-54 | 54.88% (215) | NAD dependent epimerase/dehydratase family protein |
| M. thermoresistible (build 8) | TH_3475 | - | 9e-43 | 44.86% (214) | PUTATIVE NAD dependent epimerase/dehydratase family protein |
| M. ulcerans Agy99 | MUL_0137 | - | 2e-41 | 43.46% (214) | hypothetical protein MUL_0137 |
| M. vanbaalenii PYR-1 | Mvan_4986 | - | 1e-55 | 53.67% (218) | NAD-dependent epimerase/dehydratase |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_4327|M.marinum_M -MTRVVVFGGHGKVALLLGHILADRGDQVSSVFRNPDHRDDIAATGATPV
MUL_0137|M.ulcerans_Agy99 -MTPVVVFGGHGKVALLLGHILADRGDQVSSVFRNPDHRDDIAATGATPV
MAV_1263|M.avium_104 -----MIVGGHGKVALQLSAILTQRGDAVTSLFRNPDHADDVAATGAKPV
TH_3475|M.thermoresistible__bu -MARIVIIGGHGQVAMHLSRILSERGDQVTSVFRNPNHAEEVMVTGAQPL
Mflv_1761|M.gilvum_PYR-GCK --MRVVIAGGHGKIALITSRLLSARGDSVAGLIRNPAHEDDLRVTGAEPV
Mvan_4986|M.vanbaalenii_PYR-1 --MRVVIAGGHGKIALITARLLSVRGDSVAGLIRNPDHADDLRVAGAEPV
MSMEG_2941|M.smegmatis_MC2_155 --MRVVIAGGHGKIALILERLLADRGDSVVGLIRNPEHAADVKAAGAEPI
MAB_3427|M.abscessus_ATCC_1997 MPQKVVIAGGHGQIAAHLIRLLAARGDTAVALIRNPDHAADVEAWGGLPL
:: ****::* :*: *** . .::*** * :: . *. *:
MMAR_4327|M.marinum_M QADIEGLDTAALAGLLTGHDAVVFSAGAG-GGNPARTYAVDRDAAIRVID
MUL_0137|M.ulcerans_Agy99 QADIEGLDTAALAGLLTGHDAVVFSAGAG-GGNPARTYAVDRDAATRVID
MAV_1263|M.avium_104 VADIERLDTDALAGLLAGHDAVVFSAGAG-GGNPARTYAVDRDAAIRVVD
TH_3475|M.thermoresistible__bu RADLEQLDTDTLTEVLSGHDAVVFSAGAG-GGNPTRTYAVDRDAAIRVID
Mflv_1761|M.gilvum_PYR-GCK VLDLEDTTNGVVSSHLRGADAVIFAAGAGPGSGAARKETVDRDAAVLLAD
Mvan_4986|M.vanbaalenii_PYR-1 VVDLENTTNGVVSTHLRGADAVIFAAGAGPGSGAARKETVDRDAAILLAD
MSMEG_2941|M.smegmatis_MC2_155 VLDLEKAAVDEVAGAVRGADAVVFAAGAGPGSGAARKQTVDRDAAILLAD
MAB_3427|M.abscessus_ATCC_1997 VLDLESTDADDVAGALAGADAAVFAAGAGPGSGPARKDTVDRAAAVLLAT
*:* :: : * **.:*:**** *...:*. :*** ** :
MMAR_4327|M.marinum_M AATRAGVQRFVMVSYFGAG-PNHGVSVDDSFFPYAQAKAAADAHLRA-SN
MUL_0137|M.ulcerans_Agy99 AATRAGVQRFVMISYFGAG-PNHGVSVDDSFFPYAQAKAAADAHLRA-SN
MAV_1263|M.avium_104 AAARSGVKRFVMVSYFGAG-PDHGVPQDDPFFPYAESKAAADAHLRA-SD
TH_3475|M.thermoresistible__bu AAGKAGVSRFIMVSYFGAG-PDHGVPEDSSFFPYAESKAAADAHLRA-SE
Mflv_1761|M.gilvum_PYR-GCK AAEAAGVQRYLMISAMGADANTPNDAADDVFVAYLRAKGAADDLIRSRTE
Mvan_4986|M.vanbaalenii_PYR-1 AAEAAGVPRYVMVSSMGADAAAPDDAGDDVFVAYLRAKGAADDVIRARTS
MSMEG_2941|M.smegmatis_MC2_155 AAEAAGVDRYVMVSALAADDRSLDESYDEVFRVYMQAKSEADANVRARSG
MAB_3427|M.abscessus_ATCC_1997 AAEQAQVHRFIQISAFGAGEPAPTEG-DESWIAYVIAKTAAEEDLRTRDR
** : * *:: :* :.*. *. : * :* *: :*:
MMAR_4327|M.marinum_M LDWTVLGPSRLTMEPATGRISIGTGKSGKATVSRADVARVIAATLNDDST
MUL_0137|M.ulcerans_Agy99 LDWTVLGPSRLTMEPATGRISIGTGKSGKATVSRVDVARVIAATLNDDST
MAV_1263|M.avium_104 LDWTILGPGRLTLDPPTGRIAVGRGK---GEVSRADVASVAAAALADDST
TH_3475|M.thermoresistible__bu LDWTILGPGRLTEEPATGKITVGPEAAD-SEVPREDVALVAAACLADDST
Mflv_1761|M.gilvum_PYR-GCK LYATIVRPGQLTDDPGTGKVAIAEHTG-RGKIPRADVAAVLVAVLDAPQT
Mvan_4986|M.vanbaalenii_PYR-1 LLATIVRPGLLTDDEPTGRVRIAEHTG-RGSIPRADVAAVLVAVLDTPDT
MSMEG_2941|M.smegmatis_MC2_155 LRTTIVRPGGLTNDAGTGLVRIAESTG-RGTVPREDVARVLVAVLDAPQT
MAB_3427|M.abscessus_ATCC_1997 LDWTILRPGLLTDTAPTGSVTLSASKIERGSVPRADVAAVVAELLGAPNT
* *:: *. ** ** : :. . :.* *** * . * .*
MMAR_4327|M.marinum_M IGRTIDFNNGDVPIATALAD----
MUL_0137|M.ulcerans_Agy99 IGRTIDFNNGDVPIATALAD----
MAV_1263|M.avium_104 IRRTIDFNNGDVPIAVALAG----
TH_3475|M.thermoresistible__bu VRRTIEFNKGDTSIAEAVRAR---
Mflv_1761|M.gilvum_PYR-GCK AGQTFELISGDTDIEVAVASLAVG
Mvan_4986|M.vanbaalenii_PYR-1 AGRTFEVISGDAAVADAVAALT--
MSMEG_2941|M.smegmatis_MC2_155 AGRTFELISGATPIDDALV-----
MAB_3427|M.abscessus_ATCC_1997 VHEILFLTEGTAPIASAVAGL---
. : . .* . : *: