For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VASDYIAVHNGVLHITIATAAAGTSLDFTGVDAAVQVLRDLPAEVGAILLTSSGPNFCAGGNVRDFASAE DRSAKLHDLAGRLHAFVRAVDAADRPVVASVHGWAAGGGLSLVLLADIAIGGQSTKLRAAYPGIGYSSDG GMSWALPRVVGKARAADILLTDRVVEASEALSIGLLSRVVADDEVQATAVATAEQLAAGPREAQNRIRRL LRLSATNTLDAQLDLEQTSIAEASVSPTGIEGVDAFVGKRKPVWP
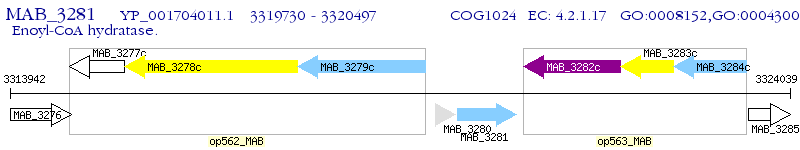
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_3281 | - | - | 100% (255) | putative enoyl-CoA hydratase/isomerase |
| M. abscessus ATCC 19977 | MAB_0905 | - | 3e-29 | 31.50% (254) | putative enoyl-CoA hydratase/isomerase |
| M. abscessus ATCC 19977 | MAB_4570c | - | 1e-19 | 25.95% (262) | enoyl-CoA hydratase/isomerase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1099c | echA8 | 1e-22 | 33.19% (226) | enoyl-CoA hydratase |
| M. gilvum PYR-GCK | Mflv_2902 | - | 4e-39 | 40.72% (221) | enoyl-CoA hydratase/isomerase |
| M. tuberculosis H37Rv | Rv1070c | echA8 | 1e-22 | 33.19% (226) | enoyl-CoA hydratase |
| M. leprae Br4923 | MLBr_02402 | - | 1e-23 | 33.33% (216) | enoyl-CoA hydratase |
| M. marinum M | MMAR_4309 | echA10 | 7e-22 | 30.62% (258) | enoyl-CoA hydratase EchA10 |
| M. avium 104 | MAV_5086 | - | 2e-27 | 36.15% (213) | enoyl-CoA hydratase |
| M. smegmatis MC2 155 | MSMEG_4299 | - | 2e-36 | 38.05% (205) | enoyl-CoA hydratase/isomerase |
| M. thermoresistible (build 8) | TH_3232 | - | 1e-36 | 37.74% (212) | PUTATIVE Enoyl-CoA hydratase |
| M. ulcerans Agy99 | MUL_0985 | echA10 | 1e-22 | 31.01% (258) | enoyl-CoA hydratase |
| M. vanbaalenii PYR-1 | Mvan_3603 | - | 4e-38 | 39.05% (210) | enoyl-CoA hydratase/isomerase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1099c|M.bovis_AF2122/97 ---------MTYETILVERDQRVGIITLNRPQALNALNSQVMNEVTSAAT
Rv1070c|M.tuberculosis_H37Rv ---------MTYETILVERDQRVGIITLNRPQALNALNSQVMNEVTSAAT
MLBr_02402|M.leprae_Br4923 ---------MKYDTILVDGYQRVGIITLNRPQALNALNSQMMNEITNAAK
Mflv_2902|M.gilvum_PYR-GCK --------MTDYQTITLEQSGPVARITLNRPEAANGMNGTMTRELADAAA
Mvan_3603|M.vanbaalenii_PYR-1 --------MTEYQTITFEQAGVVARITLNRPDAANGMNGTMTRELADAAA
TH_3232|M.thermoresistible__bu --------VTEYQTLTFEQSGPVTRIVLNRPDAANGMNATMTAELADAAR
MSMEG_4299|M.smegmatis_MC2_155 ------MTEISYEAIAFEQSGPITRITLNRPDAANGMNDTMTRELADAAR
MAV_5086|M.avium_104 -----MTTSLDAPVLREVDADGVARVRLNRPQTSNGLNVETLKALHDAVL
MMAR_4309|M.marinum_M MPDTRIETLAPVAGLEVKLRGGVLSVTIDRPESLNSLTPEVMAGIADAME
MUL_0985|M.ulcerans_Agy99 MPDTRIETLAPVAGLEVKLRGGVLSVTIDRPESLNSLTPEVMAGIADAME
MAB_3281|M.abscessus_ATCC_1997 -----------MASDYIAVHNGVLHITIATAAAGTSLDFTGVDAAVQVLR
: : : . : ..: ..
Mb1099c|M.bovis_AF2122/97 ELDDDPDIGAIIITGSAKAFAAGADIKEMADLT-----FADAFTADFFAT
Rv1070c|M.tuberculosis_H37Rv ELDDDPDIGAIIITGSAKAFAAGADIKEMADLT-----FADAFTADFFAT
MLBr_02402|M.leprae_Br4923 ELDIDPDVGAILITGSPKVFAAGADIKEMASLT-----FTDAFDADFFSA
Mflv_2902|M.gilvum_PYR-GCK RCDT-PATKVVVLTGAGRFFCAGGDLKDFASAPSRG-QHVKGVADDLHRA
Mvan_3603|M.vanbaalenii_PYR-1 RCDT-AATKVVVITGAGRFFCAGGDLKDFASAPSRG-RHVKGVADDLHRA
TH_3232|M.thermoresistible__bu RCDN-AATKVVVLTGAGRFFSAGGDLKDFADAADRS-RHIKGVADDLHRA
MSMEG_4299|M.smegmatis_MC2_155 RADT-SGAKVVVLTGSGRFFSAGGDLKAFSTAPSRG-AFIKGVADDLHRA
MAV_5086|M.avium_104 TCHADPTVKVVLLSGAGRNFCAGGDVHTFESKGADLPDYLREATAWLQLA
MMAR_4309|M.marinum_M AAATDSRVRVVRLGGTGRGFSSGAGMSAQDMGRKTS---TSSLT-EINRA
MUL_0985|M.ulcerans_Agy99 AAATDSRVRVVRLGGTGRGFSSGAGMSAQDMGRKTS---TSSLT-EINRA
MAB_3281|M.abscessus_ATCC_1997 DLPAEVGAILLTSSGPNFCAGGNVRDFASAEDRSAK---LHDLAGRLHAF
: *. .. :
Mb1099c|M.bovis_AF2122/97 WGKLAAVRTPTIAAVAGYALGGG-CELAMMCDVLIAADTAKFGQPEIKLG
Rv1070c|M.tuberculosis_H37Rv WGKLAAVRTPTIAAVAGYALGGG-CELAMMCDVLIAADTAKFGQPEIKLG
MLBr_02402|M.leprae_Br4923 WGKLAAVRTPMIAAVAGYALGGG-CELAMMCDLLIAADTAKFGQPEIKLG
Mflv_2902|M.gilvum_PYR-GCK ISSFARMDAVVITVVNGTAAGAG-FSLAVSGDLVLAAESASFTMAYTKVG
Mvan_3603|M.vanbaalenii_PYR-1 ISTFARMDAVVITAVNGTAAGAG-FSLAVSGDLVLAAESASFTMAYTRVG
TH_3232|M.thermoresistible__bu VSTFARMNAVVITAVNGTAAGAG-FSLAVTGDLVLAAESASFTMAYTRVG
MSMEG_4299|M.smegmatis_MC2_155 ISSFARMDAVLITAVNGTAAGAG-FSIAVTGDLVLAAESASFTMAYTKVG
MAV_5086|M.avium_104 TAALIQLRAPVVTAVQGFAAGGGGLGLVCASDVVLAGRSAKFFSGAVRVG
MMAR_4309|M.marinum_M VRAITALPHPVVAVVHGPAAGVG-VSLALACDLVLASESAYFLLAFTKIG
MUL_0985|M.ulcerans_Agy99 VRAITALPHPVVAVVHGPAAGVG-VSLALACDLVLASESAYFLLAFTKIG
MAB_3281|M.abscessus_ATCC_1997 VRAVDAADRPVVASVHGWAAGGG-LSLVLLADIAIGGQSTKLRAAYPGIG
. :: * * * * * :. *: :.. :: : :*
Mb1099c|M.bovis_AF2122/97 VLPGMGGSQRLTRAIGKAKAMDLILTGRTMDAAEAERSGLVSRVVPADDL
Rv1070c|M.tuberculosis_H37Rv VLPGMGGSQRLTRAIGKAKAMDLILTGRTMDAAEAERSGLVSRVVPADDL
MLBr_02402|M.leprae_Br4923 VLPGMGGSQRLTRAIGKAKAMDLILTGRTIDAAEAERSGLVSRVVLADDL
Mflv_2902|M.gilvum_PYR-GCK LSPDGSASYYLPRLIGIAKTKQLMLTNRVLSADEASRWGLVTEVVADDEL
Mvan_3603|M.vanbaalenii_PYR-1 LSPDGSASYHLPRLIGIAKTKQLMLTNRTLSAQEALQWGLVGEVVADDRL
TH_3232|M.thermoresistible__bu LSPDGAASYYLPRLIGITKTKELMLTNRTLSAQEAATWGLVTEVVADDQL
MSMEG_4299|M.smegmatis_MC2_155 LSPDGSASYYLPRLVGMRRAQELMLTNRTLSATEAYEWGLVTEVVPDADL
MAV_5086|M.avium_104 MAPDGGSSVTLTQLVGLRQALRILLTNPTLGAEEAAAIGLITDVVDDDAL
MMAR_4309|M.marinum_M LMPDGGASALIAAAIGRIRAMRMALLPERLPATEALSYGLVSAVYPVADF
MUL_0985|M.ulcerans_Agy99 LMPDGGASALIAAVIGRIRAMRMALLPERLPATEALSYGLVSAVYPVADF
MAB_3281|M.abscessus_ATCC_1997 YSSDGGMSWALPRVVGKARAADILLTDRVVEASEALSIGLLSRVVADDEV
.. . * :. :* :: : * : * ** **: * .
Mb1099c|M.bovis_AF2122/97 LTEARATATTISQMSASAARMAKEAVNRAFESSLSEGLLYERRLFHSAFA
Rv1070c|M.tuberculosis_H37Rv LTEARATATTISQMSASAARMAKEAVNRAFESSLSEGLLYERRLFHSAFA
MLBr_02402|M.leprae_Br4923 LPEAKAVATTISQMSRSATRMAKEAVNRSFESTLAEGLLHERRLFHSTFV
Mflv_2902|M.gilvum_PYR-GCK AARADALAGQMAATAAGSNGGVKALLLDTFSNGLEQQMELEGRLIAQRAE
Mvan_3603|M.vanbaalenii_PYR-1 QARADELAEQMAETSAGSNGGVKALLSDTFANGLEQQMELEGRLIAARAE
TH_3232|M.thermoresistible__bu AARADQLAEQLAQTAGGSNGGVKNLLLSTFANGLEEQMEAEGRLIARLAD
MSMEG_4299|M.smegmatis_MC2_155 ADRATKLAEQVAAGSAGSNGAVKSLLLSSLKNSLEEQMEAEGRFIAARAD
MAV_5086|M.avium_104 TEEAERIAKTLAAMPPKALSATKRLVWSGVGVPVESRLAEEARIVSELSG
MMAR_4309|M.marinum_M QNEVDAVISRLLSGPAVAFAKTKDSINAATLTELDRTLDREFQGQSALLR
MUL_0985|M.ulcerans_Agy99 QNEVDAVISRLLSGPAVAFAKTKNSINAATLTELDRTLDREFQGQSALLR
MAB_3281|M.abscessus_ATCC_1997 QATAVATAEQLAAGPREAQNRIRRLLRLSATNTLDAQLDLEQTSIAEASV
. : . : : : : : *
Mb1099c|M.bovis_AF2122/97 TEDQSEGMAAFIEKRAPQFTHR---
Rv1070c|M.tuberculosis_H37Rv TEDQSEGMAAFIEKRAPQFTHR---
MLBr_02402|M.leprae_Br4923 TDDQSEGMAAFIEKRAPQFTHR---
Mflv_2902|M.gilvum_PYR-GCK SADGREGVDAFLAKRKPEFSPGG--
Mvan_3603|M.vanbaalenii_PYR-1 SADGREGVDAFLAKRRPKFGAQSAG
TH_3232|M.thermoresistible__bu SADGREGVDAFLAKRRPAFG-----
MSMEG_4299|M.smegmatis_MC2_155 SADGREGVDAFLGKRRPEFA-----
MAV_5086|M.avium_104 TADALEGLRAVIERRPPRFTGA---
MMAR_4309|M.marinum_M SPDFAEGARAFQQRRTPTFTDS---
MUL_0985|M.ulcerans_Agy99 SPDFAEGARAFQQRRTPTFTDS---
MAB_3281|M.abscessus_ATCC_1997 SPTGIEGVDAFVGKRKPVWP-----
: ** *. :* * :