For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTNGLRPSGSSPTADTPSYDLVVIGSGPGGQKAAIAAAKLGKSVAVVERDNMLGGVCTNTGTIPSKTLRE AVLYLTGMNQRELYGASYRVKANITPEDLLARTEHVIRKEIEVVRAQLQRNRIDLISGIGRFADEHTVVV EEPSRGERTTLHADYVVIATGTNPARPDGVAFDEKRVLDSDGILNLRFIPGSMVVVGAGVIGIEYASMFA ALGTKVTVVEKRDTMLDFCDPEVVEALRFHLRDLAVTFRFGEEVTTVEVNDTGTMTTLASGKQIPADTVM YSAGRQGQTDQLDLPKAGLEADRRGRIWVDANFQTKVDHIYAVGDVIGFPALAATSMDQGRLAAYHAFGE PSKGMTDLQPIGIYSIPEVSYVGATEVELTKNAIPYEVGVSRYRELARGQIAGDSYGMLKLLVSTEDLRL LGIHIFGSDATDLVHIGQAVMGCGGTVEYLVDAVFNYPTFSEAYKVAALDVMNKIRALNQFKA
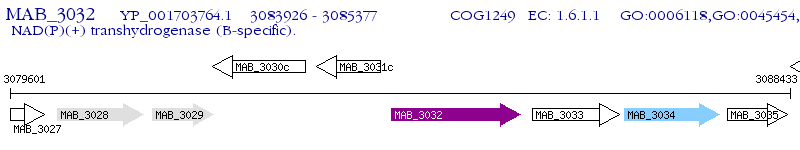
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_3032 | - | - | 100% (483) | soluble pyridine nucleotide transhydrogenase |
| M. abscessus ATCC 19977 | MAB_4127c | - | 1e-48 | 32.03% (462) | dihydrolipoamide dehydrogenase |
| M. abscessus ATCC 19977 | MAB_0894c | - | 2e-41 | 28.73% (456) | dihydrolipoamide dehydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2732 | sthA | 0.0 | 83.41% (464) | soluble pyridine nucleotide transhydrogenase |
| M. gilvum PYR-GCK | Mflv_0114 | - | 2e-49 | 33.04% (457) | dihydrolipoamide dehydrogenase |
| M. tuberculosis H37Rv | Rv2713 | sthA | 0.0 | 83.41% (464) | soluble pyridine nucleotide transhydrogenase |
| M. leprae Br4923 | MLBr_02387 | lpd | 1e-45 | 30.91% (453) | dihydrolipoamide dehydrogenase |
| M. marinum M | MMAR_2000 | sthA | 0.0 | 82.76% (464) | soluble pyridine nucleotide transhydrogenase SthA |
| M. avium 104 | MAV_3606 | - | 0.0 | 84.48% (464) | soluble pyridine nucleotide transhydrogenase |
| M. smegmatis MC2 155 | MSMEG_2748 | sthA | 0.0 | 85.13% (464) | soluble pyridine nucleotide transhydrogenase |
| M. thermoresistible (build 8) | TH_3289 | - | 0.0 | 77.63% (465) | PUTATIVE acyl-CoA dehydrogenase, C- domain protein |
| M. ulcerans Agy99 | MUL_2214 | lpd | 4e-45 | 31.28% (454) | dihydrolipoamide dehydrogenase |
| M. vanbaalenii PYR-1 | Mvan_2448 | - | 0.0 | 84.73% (465) | soluble pyridine nucleotide transhydrogenase |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_2748|M.smegmatis_MC2_155 ------------MGSMLEYDLVVIGSGPGGQKAAIAAAKLGKSVAVVERG
Mvan_2448|M.vanbaalenii_PYR-1 ------------MASKLEYDLVVIGSGPGGQKAAIAAAKLGKTVAVVERG
Mb2732|M.bovis_AF2122/97 ---------------MREYDIVVIGSGPGGQKAAIASAKLGKSVAIVERG
Rv2713|M.tuberculosis_H37Rv ---------------MREYDIVVIGSGPGGQKAAIASAKLGKSVAIVERG
MMAR_2000|M.marinum_M ------------MGSMREYDMVVIGSGPGGQKAAIASAKLGKSVAIIERG
MAV_3606|M.avium_104 ------------MSSPQEYDMVVIGSGPGGQKAAIASAKLGKSVAVVERG
MAB_3032|M.abscessus_ATCC_1997 MTNGLRPSGSSPTADTPSYDLVVIGSGPGGQKAAIAAAKLGKSVAVVERD
TH_3289|M.thermoresistible__bu ------------MG--AKYDLIVIGSGPGGQKAAIAAAKLGKTVAVVERG
Mflv_0114|M.gilvum_PYR-GCK ---------------MTHYDVVVLGAGPGGYVAAIRAAQLGLSTAIVEP-
MUL_2214|M.ulcerans_Agy99 ---------------MTHYDVVVLGAGPAGYVAAIRAAQLGLNTAIVEP-
MLBr_02387|M.leprae_Br4923 ---------------MTHYDVVVLGAGPGGYVAAIRAAQLGLSTAVVEP-
**::*:*:**.* *** :*:** ..*::*
MSMEG_2748|M.smegmatis_MC2_155 QMLGGVCVNTGTIPSKTLREAVVYLTGMNQRELYGASYRVKEKITPADLM
Mvan_2448|M.vanbaalenii_PYR-1 RMLGGVCVNTGTIPSKTLREAVVYLTGMSQRELYGTSYRVKEKITPADLL
Mb2732|M.bovis_AF2122/97 RMLGGVCVNTGTIPSKTLREAVLYLTGMNQRELYGASYRVKDRITPADLL
Rv2713|M.tuberculosis_H37Rv RMLGGVCVNTGTIPSKTLREAVLYLTGMNQRELYGASYRVKDRITPADLL
MMAR_2000|M.marinum_M RMLGGVCVNTGTIPSKTLREAVLYLTGMNQRELYGASYRVKDRITPADLL
MAV_3606|M.avium_104 QMLGGVCVNTGTIPSKTLREAVLYLTGMSQRELYGASYRVKEKITPADLL
MAB_3032|M.abscessus_ATCC_1997 NMLGGVCTNTGTIPSKTLREAVLYLTGMNQRELYGASYRVKANITPEDLL
TH_3289|M.thermoresistible__bu RMLGGVCINTGTIPSKTLREAVLYLTGMNQRELYGAGYRVKDHITPADLL
Mflv_0114|M.gilvum_PYR-GCK KYWGGVCLNVGCIPSKALLRN-AELAHVFHKDAKTFGISGDVSFDYGAAF
MUL_2214|M.ulcerans_Agy99 KYWGGVCLNVGCIPSKALLRN-AELAHIFTKDAKAFGISGEATFDYGVAF
MLBr_02387|M.leprae_Br4923 KYWGGICLNVGCIPSKVLLHN-AELAHIFTKEAKTFGISGDASFDYGIAY
. **:* *.* ****.* . *: : :: . . :
MSMEG_2748|M.smegmatis_MC2_155 ARTQHVISREQDVVRSQLMRNRVDIISGHGRFVDPHTVLVEEPSRGE--R
Mvan_2448|M.vanbaalenii_PYR-1 ARTTHVISREQDVVRSQLMRNRVDLVQGHGRFLDAHTVLVEEPHRGE--R
Mb2732|M.bovis_AF2122/97 ARTQHVIGKEVDVVRNQLMRNRVDLIVGHGRFIDPHTILVEDQARRE--K
Rv2713|M.tuberculosis_H37Rv ARTQHVIGKEVDVVRNQLMRNRVDLIVGHGRFIDPHTILVEDQARRE--K
MMAR_2000|M.marinum_M ARTQHVIGKEVDVVRSQLMRNRVDLIVGHGRFVDPHSIVVEDQTHRE--K
MAV_3606|M.avium_104 ARTQHVIGKEIDVVRNQLMRNRIDLLIGHGRFVDPHTIEVEDPSRRE--K
MAB_3032|M.abscessus_ATCC_1997 ARTEHVIRKEIEVVRAQLQRNRIDLISGIGRFADEHTVVVEEPSRGE--R
TH_3289|M.thermoresistible__bu SRTRHVIEREIEVVRDQLTRNNVDLHTGHGRFRDPHTIVVEDAARAE--L
Mflv_0114|M.gilvum_PYR-GCK DRSRKVADGRVAGVHFLMKKNKITEVHGYGRFTGPNSIEVEPTGDGEEGP
MUL_2214|M.ulcerans_Agy99 DRSRKVAEGRVAGVHFLMKKNKITEIHGYGKFTDANTLSVDLN-DG--GT
MLBr_02387|M.leprae_Br4923 DRSRKVSEGRVAGVHFLMKKNKITEIHGYGRFTDANTLSVELSEGVPETP
*: :* . *: : :*.: * *:* . ::: *:
MSMEG_2748|M.smegmatis_MC2_155 TMVGGEYIVIATGTKPARPAGVEFDERRVLDSDGILDLVSLPASMVVVGA
Mvan_2448|M.vanbaalenii_PYR-1 TTLTGEHIVIATGTKPARPAGVEFDEERVLDSDGILDLKTLPTSMVVVGA
Mb2732|M.bovis_AF2122/97 TTVTGDYIIIATGTRPARPSGVEFDEERVLDSDGILDLKSLPSSMVVVGA
Rv2713|M.tuberculosis_H37Rv TTVTGDYIIIATGTRPARPSGVEFDEERVLDSDGILDLKSLPSSMVVVGA
MMAR_2000|M.marinum_M TTVTGDYIVIATGTRPARPSGVEFDEDKVLDSDGILDLKSLPASMVVVGA
MAV_3606|M.avium_104 ITISGKYVVIATGTRPARPSGVEFDEDRVLDSDGILDLKSLPTSMVVVGA
MAB_3032|M.abscessus_ATCC_1997 TTLHADYVVIATGTNPARPDGVAFDEKRVLDSDGILNLRFIPGSMVVVGA
TH_3289|M.thermoresistible__bu TTLTGDYIVLAPGTSPARPQGVAFDERRVLDSDGILDLEALPGSMVVVGA
Mflv_0114|M.gilvum_PYR-GCK VKLEFDNAIIATGSSTRLVPGTSLSEN-VVTYEKQILTRELPGSIIIAGA
MUL_2214|M.ulcerans_Agy99 EKVTFDNAIIATGSSTRLVPGTSLSTN-VVTYEEQILTRELPKSIIIAGA
MLBr_02387|M.leprae_Br4923 LKVTFNNVIIATGSKTRLVPGTLLSTN-VITYEEQILTRELPDSIVIVGA
: . ::*.*: . *. :. *: : : :* *:::.**
MSMEG_2748|M.smegmatis_MC2_155 GVIGIEYASMFAALGTKVTVVEKRDNMLEFCDPEIVEALKFHLRDLAVTF
Mvan_2448|M.vanbaalenii_PYR-1 GVIGIEYASMFAALGTKVTVVEKRGSMLEFCDPEIVESLKFHLRDLAVTF
Mb2732|M.bovis_AF2122/97 GVIGIEYASMFAALGTKVTVVEKRDNMLDFCDPEVVEALKFHLRDLAVTF
Rv2713|M.tuberculosis_H37Rv GVIGIEYASMFAALGTKVTVVEKRDNMLDFCDPEVVEALKFHLRDLAVTF
MMAR_2000|M.marinum_M GVIGIEYASMFAALGTKVTVVEKRDNMLDFCDPEVVEALKFHLRDLAVTF
MAV_3606|M.avium_104 GVIGIEYASMFAALGTKVTVVEKRDDMLDFCDPEVVEALKFHLRDLAVTF
MAB_3032|M.abscessus_ATCC_1997 GVIGIEYASMFAALGTKVTVVEKRDTMLDFCDPEVVEALRFHLRDLAVTF
TH_3289|M.thermoresistible__bu GVIGIEYASMFAALGTRVTVVEKRASMLDFCDPEIVEALTYHLRDLAVTF
Mflv_0114|M.gilvum_PYR-GCK GAIGMEFAYVLKNYGVDVTIVEFLPRALPNEDAEVSKEIEKQYKKLGVKI
MUL_2214|M.ulcerans_Agy99 GAIGMEFGYVLKNYGVDVTIVEFLPRALPNEDAEVSKEIERQFKKLGVKI
MLBr_02387|M.leprae_Br4923 GAIGIEFGYVLKNYGVDVTIVEFLPRAMPNEDAEVSKEIEKQFKKMGIKI
*.**:*:. :: *. **:** : *.*: : : : :.:.:.:
MSMEG_2748|M.smegmatis_MC2_155 RFGEEVTAVDVGSAGTVTTLASG---KQIPADTVMYSAGRQGQTEHLDLA
Mvan_2448|M.vanbaalenii_PYR-1 RFGEEVTGVDVGAAGTVTTLASG---KQIPAETVMYSAGRQGQTDHLDLA
Mb2732|M.bovis_AF2122/97 RFGEEVTAVDVGSAGTVTTLASG---KQIPAETVMYSAGRQGQTDHLDLH
Rv2713|M.tuberculosis_H37Rv RFGEEVTAVDVGSAGTVTTLASG---KQIPAETVMYSAGRQGQTDHLDLH
MMAR_2000|M.marinum_M RFGEEVTAVDVGSAGTVTTLASG---KQIPAETVMYSAGRQGQTDHLDLA
MAV_3606|M.avium_104 RFGEEVTAVDVGSAGTVTTLASG---KQIPAETVMYSAGRQGQTDHLDLH
MAB_3032|M.abscessus_ATCC_1997 RFGEEVTTVEVNDTGTMTTLASG---KQIPADTVMYSAGRQGQTDQLDLP
TH_3289|M.thermoresistible__bu RFGEQVTGVEVSPAGTVTSLASG---KRIPADAVMYSAGRQGQTARLDLH
Mflv_0114|M.gilvum_PYR-GCK LTGTKVEKIEDDGKQVTVTVSKNDKTEELKADKVMQAIGFAPNVEGFGLD
MUL_2214|M.ulcerans_Agy99 RTGTKVESISDDGSAVTVVVSKGGKSEELKADKVLQAIGFAPNVEGYGLD
MLBr_02387|M.leprae_Br4923 LTGTKVESISDNGSHVLVAVSKDGQFQELKADKVLQAIGFAPNVDGYGLD
* :* :. . . . ::.. :.: *: *: : * :. .*
MSMEG_2748|M.smegmatis_MC2_155 NAGLGVDGRGRITVDSNFQTKVDHIYAVGDVIGFPAQAATSMDQGRLAAY
Mvan_2448|M.vanbaalenii_PYR-1 SAGLEADDRGRIFVDDNYQTKVDHIYAVGDVIGFPALAATSMDQGRLAAY
Mb2732|M.bovis_AF2122/97 NAGLEVQGRGRIFVDDRFQTKVDHIYAVGDVIGFPALAATSMEQGRLAAY
Rv2713|M.tuberculosis_H37Rv NAGLEVQGRGRIFVDDRFQTKVDHIYAVGDVIGFPALAATSMEQGRLAAY
MMAR_2000|M.marinum_M NAGLEVEGRGRIWVDDKFRTKVEHIYAVGDVIGFPALAATSMEQGRLAAY
MAV_3606|M.avium_104 NAELEADNRGRIFVDDNFQTKVPHIYAVGDVIGFPALAATSMEQGRLAAY
MAB_3032|M.abscessus_ATCC_1997 KAGLEADRRGRIWVDANFQTKVDHIYAVGDVIGFPALAATSMDQGRLAAY
TH_3289|M.thermoresistible__bu NAGLEADDRGRIAVDDNYRTAVDHIYAVGDVIGFPALASTSMEQGRVAAC
Mflv_0114|M.gilvum_PYR-GCK KAGVELTDRKAIGIDDYMRTNVPHIYAIGDVTAKLQLAHVAEAMGVVAAE
MUL_2214|M.ulcerans_Agy99 KAGVALTDRKAIGIGEYMQTSVGHIYAIGDVTGQLMLAHVAEAMGVVAAE
MLBr_02387|M.leprae_Br4923 KVGVALTADKAVDIDDYMQTNVSHIYAIGDVTGKLQLAHVAEAQGVVAAE
.. : : :. :* * ****:*** . * .: * :**
MSMEG_2748|M.smegmatis_MC2_155 HAFGEPAKGMTDLQPIG--IYSIPEVSYVGATEVDLTRNAIPYEVGVSRY
Mvan_2448|M.vanbaalenii_PYR-1 HAFGEPTQGMTELQPIG--IYSIPEVSYVGATEVELTKNAIPYEVGVSRY
Mb2732|M.bovis_AF2122/97 HAFGEPTDGITELQPIG--IYSIPEVSYVGATEVELTKSSIPYEVGVARY
Rv2713|M.tuberculosis_H37Rv HAFGEPTDGITELQPIG--IYSIPEVSYVGATEVELTKSSIPYEVGVARY
MMAR_2000|M.marinum_M HAFGEPTDGITELQPIG--IYSIPEISYVGATEVELTKNSIPYEVGVARY
MAV_3606|M.avium_104 HAFGEASDGITELQPIG--IYSIPEVSYVGATEVELTKDAIPYEVGVARY
MAB_3032|M.abscessus_ATCC_1997 HAFGEPSKGMTDLQPIG--IYSIPEVSYVGATEVELTKNAIPYEVGVSRY
TH_3289|M.thermoresistible__bu HAFGEPVDQLPELHPVG--IYAIPEISYVGSTEVELTRDAVPYETGVARY
Mflv_0114|M.gilvum_PYR-GCK TIAGADTLPFEDYRMMPRATFCQPQVASFGLTEQQAKDEGYDVVVAKFPF
MUL_2214|M.ulcerans_Agy99 TIAGAETLPLGDYRMLPRATFCQPQVASFGLTEQQARDEGYDVKVAKFPF
MLBr_02387|M.leprae_Br4923 AIAGAETLALSDYRMMPRATFCQPNVASFGLTEQQARDGGYDVVVAKFPF
* : : : : :. *::: .* ** : . .. :
MSMEG_2748|M.smegmatis_MC2_155 RELARGQIAGDSYGMLKLLVSTEDLRLLGVHIFGTSATEMVHIGQAVMGC
Mvan_2448|M.vanbaalenii_PYR-1 RELARGQIAGDSYGMLKLLVSTEDLRLLGVHIFGTSATEMVHIGQAVMGC
Mb2732|M.bovis_AF2122/97 RELARGQIAGDSYGMLKLLVSTEDLKLLGVHIFGTSATEMVHIGQAVMGC
Rv2713|M.tuberculosis_H37Rv RELARGQIAGDSYGMLKLLVSTEDLKLLGVHIFGTSATEMVHIGQAVMGC
MMAR_2000|M.marinum_M RELARGQIAGDSYGMLKLLVSTEDLTLLGVHIFGTSATEMVHIGQAIMGC
MAV_3606|M.avium_104 RELARGQIAGDSYGMLKLLVSTQDLKVLGVHIFGTSATEMVHIGQAVMGC
MAB_3032|M.abscessus_ATCC_1997 RELARGQIAGDSYGMLKLLVSTEDLRLLGIHIFGSDATDLVHIGQAVMGC
TH_3289|M.thermoresistible__bu RELARGQIAGDSYGMLKLLVSTEDRTVLGVHIVGTHATELVHIGQAVMGC
Mflv_0114|M.gilvum_PYR-GCK TANGKAHGLGDPTGFVKLIADKKHLELLGGHLIGPDVAELLPELTLAQKW
MUL_2214|M.ulcerans_Agy99 TANGKAHGLGDPSGFVKLVADGKHGELLGGHLVGHDVSELLPELTLAQKW
MLBr_02387|M.leprae_Br4923 TANAKAHGMGDPSGFVKLVADAKYGELLGGHMIGHNVSELLPELTLAQKW
.:.: **. *::**:.. : :** *:.* .::::
MSMEG_2748|M.smegmatis_MC2_155 GGTIEYLVDAVFNYPTFSEAYKVAALDVMNKLRALSRFRT
Mvan_2448|M.vanbaalenii_PYR-1 GGTVEYLVDAVFNYPTFSEAYKVAALDVMNKLRALSQFRS
Mb2732|M.bovis_AF2122/97 GGSVEYLVDAVFNYPTFSEAYKNAALDVMNKMRALNQFRR
Rv2713|M.tuberculosis_H37Rv GGSVEYLVDAVFNYPTFSEAYKNAALDVMNKMRALNQFRR
MMAR_2000|M.marinum_M GGTVEYLVEAVFNYPTFSEAYKVAALDVMNKVRALNQFRR
MAV_3606|M.avium_104 GGTVEYLVDAVFNYPTFSEAYKVAALDVMNKVRALNQFRR
MAB_3032|M.abscessus_ATCC_1997 GGTVEYLVDAVFNYPTFSEAYKVAALDVMNKIRALNQFKA
TH_3289|M.thermoresistible__bu GGTVDYLVDAVFNYPTYAEAYKVAALDVANKLRTLDRLQS
Mflv_0114|M.gilvum_PYR-GCK DLTANELARNVHTHPTLSEAMQECFHGLVGHMINF-----
MUL_2214|M.ulcerans_Agy99 DLTATELARNVHTHPTMSEALQECFHGLAGHMINF-----
MLBr_02387|M.leprae_Br4923 DLTATELVRNVHTHPTLSEALQECFHGLIGHMINF-----
. : *. *..:** :** : . .: .:: :