For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VNDLVDTTEMYLRTIYDLEEEGVVPLRARIAERLEQSGPTVSQTVARMERDGLLNVAGDRHLELTDKGRA LAVSVMRKHRLAECLLVDIIGLPWEDVHAEACRWEHVMSEDVERRLLTVLNNPTTSPFGNPIPGLSELGV PADRSESASAVRLTEIPSGSQVAVVVRRLAEHVQADTDLLSRLKDAGVVPNARVTVESSPDGVIIIIPGH ESVELPHEMAHAVMVEKV
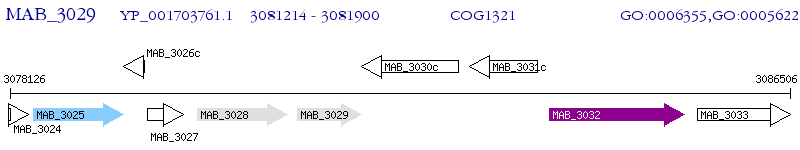
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_3029 | - | - | 100% (228) | Iron-dependent repressor IdeR |
| M. abscessus ATCC 19977 | MAB_3110 | - | 7e-20 | 31.00% (200) | iron dependent transcriptional repressor FeoA |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2730 | ideR | 1e-105 | 81.74% (230) | IRON-dependent repressor and activator IDER |
| M. gilvum PYR-GCK | Mflv_3949 | - | 1e-105 | 82.10% (229) | iron dependent repressor |
| M. tuberculosis H37Rv | Rv2711 | ideR | 1e-105 | 81.74% (230) | IRON-dependent repressor and activator IDER |
| M. leprae Br4923 | MLBr_01013 | - | 1e-104 | 82.61% (230) | iron dependent repressor |
| M. marinum M | MMAR_2002 | ideR | 1e-106 | 84.35% (230) | iron-dependent repressor and activator IdeR |
| M. avium 104 | MAV_3604 | - | 1e-107 | 83.84% (229) | iron-dependent repressor IdeR |
| M. smegmatis MC2 155 | MSMEG_2750 | - | 1e-105 | 82.61% (230) | iron-dependent repressor IdeR |
| M. thermoresistible (build 8) | TH_1487 | ideR | 1e-102 | 79.91% (229) | IRON-DEPENDENT REPRESSOR AND ACTIVATOR IDER |
| M. ulcerans Agy99 | MUL_3351 | ideR | 1e-105 | 83.91% (230) | iron-dependent repressor and activator IdeR |
| M. vanbaalenii PYR-1 | Mvan_2450 | - | 1e-105 | 82.10% (229) | iron dependent repressor |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_2002|M.marinum_M MNDLVDTTEMYLRTIYDLEEEGVTPLRARIAERLEQSGPTVSQTVSRMER
MUL_3351|M.ulcerans_Agy99 MNDLVDTTEMYLRTIYDLEEEGVTPLRARIAERLEQSGPTVSQTVSRMER
MAV_3604|M.avium_104 MNDLVDTTEMYLRTIYDLEEEGVTPLRARIAERLDQSGPTVSQTVSRMER
Mb2730|M.bovis_AF2122/97 MNELVDTTEMYLRTIYDLEEEGVTPLRARIAERLDQSGPTVSQTVSRMER
Rv2711|M.tuberculosis_H37Rv MNELVDTTEMYLRTIYDLEEEGVTPLRARIAERLDQSGPTVSQTVSRMER
MLBr_01013|M.leprae_Br4923 MNDLVDTTEMYLRTIYDLEEEGVTPLRARIAERLEQSGPTVSQTVSRMER
Mflv_3949|M.gilvum_PYR-GCK MNDLVDTTEMYLRTIYDLEEECVIPLRARIAERLDQSGPTVSQTVSRMER
Mvan_2450|M.vanbaalenii_PYR-1 MNDLVDTTEMYLRTIYDLEEEGVIPLRARIAERLDQSGPTVSQTVSRMER
MSMEG_2750|M.smegmatis_MC2_155 MNDLVDTTEMYLRTIYDLEEEGVVPLRARIAERLDQSGPTVSQTVSRMER
TH_1487|M.thermoresistible__bu MNDLVDTTEMYLRTIYDLEEEGVVPLRARIAERLEQSGPTVSQTVSRMER
MAB_3029|M.abscessus_ATCC_1997 MNDLVDTTEMYLRTIYDLEEEGVVPLRARIAERLEQSGPTVSQTVARMER
**:****************** * **********:**********:****
MMAR_2002|M.marinum_M DGLLRVAGDRHLELTDKGRALAVAVMRKHRLAERLLVDVIGLPWEEVHAE
MUL_3351|M.ulcerans_Agy99 DGLLRVAGDRHLELTDKGRALAVAVMRKHRLAERLLVDVIGLPWEEVHAE
MAV_3604|M.avium_104 DGLLHVAGDRHLELTDKGRALAVAVMRKHRLAERLLVDVIGLPWEEVHAE
Mb2730|M.bovis_AF2122/97 DGLLRVAGDRHLELTEKGRALAIAVMRKHRLAERLLVDVIGLPWEEVHAE
Rv2711|M.tuberculosis_H37Rv DGLLRVAGDRHLELTEKGRALAIAVMRKHRLAERLLVDVIGLPWEEVHAE
MLBr_01013|M.leprae_Br4923 DGLLRVAGNRHLELTTKGRAMAIAVMRKHRLAERLLVDVIGLPWEEVHAE
Mflv_3949|M.gilvum_PYR-GCK DGLLHVAGDRHLELTDKGRALAVAVMRKHRLAERLLVDVIGLPWEEVHAE
Mvan_2450|M.vanbaalenii_PYR-1 DGLLHVAGDRHLELTEKGRALAVAVMRKHRLAERLLVDVIGLPWEEVHAE
MSMEG_2750|M.smegmatis_MC2_155 DGLLHVAGDRHLELTDKGRALAVAVMRKHRLAERLLVDVIGLPWEDVHAE
TH_1487|M.thermoresistible__bu DGLLQVAGDRHLELTEKGRALAISVMRKHRLAERLLVDVIGLPWEEVHAE
MAB_3029|M.abscessus_ATCC_1997 DGLLNVAGDRHLELTDKGRALAVSVMRKHRLAECLLVDIIGLPWEDVHAE
****.***:****** ****:*::********* ****:******:****
MMAR_2002|M.marinum_M ACRWEHVMSEDVERRLVKVLNNPTTSPFGNPIPGLLDLGVGPESDSYEAN
MUL_3351|M.ulcerans_Agy99 ACRWEHVMSEDVERRLVKVLNNPTTSPFGNPIPGLLDLGVGPESDSYEAN
MAV_3604|M.avium_104 ACRWEHVMSEDVERRLVKVLNNPTTSPFGNPIPGLLDLGVGPESGAEEAN
Mb2730|M.bovis_AF2122/97 ACRWEHVMSEDVERRLVKVLNNPTTSPFGNPIPGLVELGVGPEPGADDAN
Rv2711|M.tuberculosis_H37Rv ACRWEHVMSEDVERRLVKVLNNPTTSPFGNPIPGLVELGVGPEPGADDAN
MLBr_01013|M.leprae_Br4923 ACRWEHVMSEDVERRLIKVLNNPTTSPFGNPIPGLLDLGAGPDASAANAK
Mflv_3949|M.gilvum_PYR-GCK ACRWEHVMSVDVERRLVQVLNNPTTSPFGNPIPGLSELGVGDESG-EAMS
Mvan_2450|M.vanbaalenii_PYR-1 ACRWEHVMSVDVERRLVQVLNNPTTSPFGNPIPGLSELGVGDEIN-ETFS
MSMEG_2750|M.smegmatis_MC2_155 ACRWEHVMSEEVERRLVQVLDNPTTSPFGNPIPGLTELGVTPGVNTEDVS
TH_1487|M.thermoresistible__bu ACRWEHVMSEDVERRLVKVLDNPTTSPFGNPIPGLSELGVVSTEP-EDMN
MAB_3029|M.abscessus_ATCC_1997 ACRWEHVMSEDVERRLLTVLNNPTTSPFGNPIPGLSELGVPADRS-ESAS
********* :*****: **:************** :**. .
MMAR_2002|M.marinum_M LVRLTELPSGSPVAVVVRQLTEHVQGDIDLISRLKDAGVVPNARVTVETS
MUL_3351|M.ulcerans_Agy99 LVRLTELPSGSPVAVVVRQLTEHVQGDIDLISRLKDAGVVPNARVTVETS
MAV_3604|M.avium_104 LVRLTELPSGAPVAVVVRQLTEHVQGDIDLISRLKDAGVVPNARVTVETG
Mb2730|M.bovis_AF2122/97 LVRLTELPAGSPVAVVVRQLTEHVQGDIDLITRLKDAGVVPNARVTVETT
Rv2711|M.tuberculosis_H37Rv LVRLTELPAGSPVAVVVRQLTEHVQGDIDLITRLKDAGVVPNARVTVETT
MLBr_01013|M.leprae_Br4923 LVRLTELPSGSPVAVVVRQLTEHVQDDIDLITRLKDTGVVPNARVTVETS
Mflv_3949|M.gilvum_PYR-GCK LVRLTELPAGMPVAVVVRQLTEHVQGDVELIARLKDAGVVPNARVTVENN
Mvan_2450|M.vanbaalenii_PYR-1 MVRLTELPAGMPVAVVVRQLTEHVQGDAELIARLKDAGVVPNARVTVEVK
MSMEG_2750|M.smegmatis_MC2_155 LVRLTELPVGMPVAVVVRQLTEHVQGDTDLIGRLKEAGVVPNARVTVEAN
TH_1487|M.thermoresistible__bu LVRLTELPAGTPVPVVVRQLTEHVQGDVELITRLKEAGVVPNARVTVQVN
MAB_3029|M.abscessus_ATCC_1997 AVRLTEIPSGSQVAVVVRRLAEHVQADTDLLSRLKDAGVVPNARVTVESS
*****:* * *.****:*:**** * :*: ***::**********:
MMAR_2002|M.marinum_M PAGGVTILIPGHENVTLPHAMAHAVKVEKV
MUL_3351|M.ulcerans_Agy99 PAGGVTILIPGHENVTLPHAMAYAVKVEKV
MAV_3604|M.avium_104 PAG-VTIVIPGHENVTLPHEMAHAVKVEKV
Mb2730|M.bovis_AF2122/97 PGGGVTIVIPGHENVTLPHEMAHAVKVEKV
Rv2711|M.tuberculosis_H37Rv PGGGVTIVIPGHENVTLPHEMAHAVKVEKV
MLBr_01013|M.leprae_Br4923 PAGNVIIIIPGHENVTLPHEMAHAVKVEKV
Mflv_3949|M.gilvum_PYR-GCK DHGGVMIVIPGHEQVELPHEMAHAVRVEKV
Mvan_2450|M.vanbaalenii_PYR-1 DHGGVMIVIPGHEQVELPHEMAHAVKVEKV
MSMEG_2750|M.smegmatis_MC2_155 NNGGVMIVIPGHEQVELPHHMAHAVKVEKV
TH_1487|M.thermoresistible__bu ENKSVSIIIPGHEQVDLPYQMAHAVKVEKV
MAB_3029|M.abscessus_ATCC_1997 PDG-VIIIIPGHESVELPHEMAHAVMVEKV
* *:*****.* **: **:** ****