For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SSEGVGDGAGVNLDLSLRFFSELTGVSDLRHGQFGVGNFFLPNDPASHAEAEQVIEQALRAQGFTILLSR DVPVNNDAIRPAAIKYQLPIRQWVFRSDSERKTHEALMAIESVAYTRPELLGLYPLSLSTRTQVLKGRLN SNEVVPYFSDLSDPRMEVHSMFFHTRFSTNTAPNATMAQPFRLMAHNGELNTDKKNRLSENAIARARNRA IIRPVGQSDSCRLDQTLQSRVLEDDLDLVTAVVAMMPPAWENDTTLSPRVTAMLEYFSLYEEKNDGPAAL IFGDGNIIGARLDRLGLRPLRTVETAEYLGVMSEAGQIYFEPDSVLERGRIEAGGMRYYDHSEGRSYNTV EALEMLAARHDYPAMLAQARVNIADLPHVPPAGQGSPARYDGDLDRHQRYVAYSLNQESFKFLMDPMLAT GQEKISAMGYGAAINALSNQEGGVAKYFSQRFAQVTNPPLDSIREADGMTLRVALGAKPNSGAQPAPQIV VPSPILTHLDMLKIREQGRTPVRRFGMRYRVDLTDAVSNAERIVRAIDDLCDAVEEFARETGGIAVISDR HVSSERAAMPLIIVISAINQRLIEEGLRLRVSLIAESGQLCSSHHVAAVLGFGASAVYPLAVQMRAEEKY GPDVDDAVNAFKHFAKAAEKSLMKTMGRVGLCTVESYIGGEFFEPNYLDTSDPVLRKYFPNVVSPVAGVG FSTLAAAVADWHARALAVTGEKDVPLLGLFKERAEGAGHSYGTTAVRGFVDMTEESISFGDEARPENPDM ADPTYLRLLPLHQLEGAFGLDDEAYRNNSFDELPTRAIDSFDITPGYRNFVRAMNSERTRRPAALRDVLA LPADVNFVRTADEFRREMGQFARLGNNDFQVRGLSCQQTDDVFVLRLAHSPERLAALASSLRTRFGREIT HHEIVGDELRITATGQAADYLAQIRTAPESIPLSSVQPASEITPTLTSGAMSHGALVAPAHEAVAHGTNL VGGLSNCGEGGEHISRYGTIRGSRIKQFASGRFGVWAGYLADPMLQELEIKIGQGAKPGEGGQLPAPKVT VQIAAARGGTPGVELVSPPPHHDTYSIEDLAQLIHDCKAARVRVIVKLVSSEGIGTIAVGVAKAGADVIN VAGNTGGTGAAAVTSLKYAGRSAEIGVAEVHQALCANGIRQKVVLRCSGAHQTASDVVKSALLGADSFEF GTTALMMLKCVMAKNCNIKCPAGLTTNAEVFEGDPRALAQYLLNIAHEVREILAALGMRTLREARGRSDL LHLLDHPSSIGTLDLRRMLAVAEEFVVENPVYMEKDYSVDDAFAAQFDARGAVLKPVTLTNQNKSVGGQL AIDIERSLNYQNIEGPAVATDERGRRYLLPESIAITTTGSAGQSYGVFCNDGMVLTHTGTCNDGVGKSAC GGTITVRSPGGGSSEPGGNVLIGNFALFGASGGRLFVQGQAGDRFAVRNSGATAVVEGTGEFLCEYMTNG AVLNIGDFGKGVANGMSGGFLYQYDPHGQLPSKVSHDSVLVLPITDAPFHEAAAHILLQWHVAATGSTKG QALLDDWQSARDHMVYTMSRALLQYQDSDAILQGKTRKELLDELTAALAGYQVHKFKLSYRDRRDVVGGT VPAYGDTDTEGMYALLNTYTVLNMAQQLALSRMPNVTDVTDPRIGKAVRNLVLTEDFFLIQKLQKYAREA IDGYSDEDLAVLIADKRLTDYKDALSQRNVLSMDSPGTYGWILHQSAKNIDKIGRLPSFEELFAHRALPA VALSGPSLQTT*
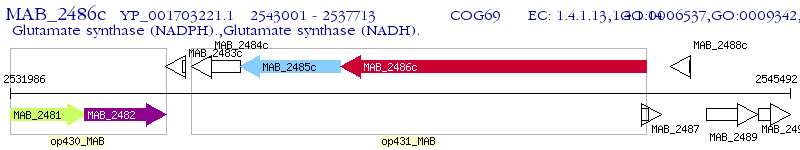
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_2486c | - | - | 100% (1762) | glutamate synthase, large subunit |
| M. abscessus ATCC 19977 | MAB_0091 | - | e-126 | 42.99% (649) | putative ferredoxin-dependent glutamate synthase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3889c | gltB | 1e-128 | 41.48% (704) | putative ferredoxin-dependent glutamate synthase |
| M. gilvum PYR-GCK | Mflv_1114 | - | 1e-130 | 42.70% (651) | glutamate synthase (ferredoxin) |
| M. tuberculosis H37Rv | Rv3859c | gltB | 1e-128 | 41.48% (704) | ferredoxin-dependent glutamate synthase |
| M. leprae Br4923 | MLBr_00061 | gltB | 1e-124 | 42.48% (652) | putative ferredoxin-dependent glutamate synthase |
| M. marinum M | MMAR_5413 | gltB | 1e-129 | 41.62% (704) | ferredoxin-dependent glutamate synthase |
| M. avium 104 | MAV_0166 | - | 1e-128 | 42.66% (647) | ferredoxin-dependent glutamate synthase 1 |
| M. smegmatis MC2 155 | MSMEG_6459 | - | 1e-128 | 41.37% (684) | ferredoxin-dependent glutamate synthase 1 |
| M. thermoresistible (build 8) | TH_2102 | gltB | 1e-129 | 43.21% (648) | PROBABLE FERREDOXIN-DEPENDENT GLUTAMATE SYNTHASE |
| M. ulcerans Agy99 | MUL_5037 | gltB | 1e-128 | 41.48% (704) | ferredoxin-dependent glutamate synthase |
| M. vanbaalenii PYR-1 | Mvan_5698 | - | 1e-130 | 42.77% (650) | glutamate synthase (ferredoxin) |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3889c|M.bovis_AF2122/97 -------------MTPKRVGLYNPAFEHDSCGVAMVVDMHGRRSRDIVDK
Rv3859c|M.tuberculosis_H37Rv -------------MTPKRVGLYNPAFEHDSCGVAMVVDMHGRRSRDIVDK
MAV_0166|M.avium_104 -----------MHMTPKRAGLYNPAFEHDACGVAMVVDMHGRRSRDIVDK
MMAR_5413|M.marinum_M -------------MTPKRVGLYNPAFEHDSCGVAMVVDMHGRRSRDIVDK
MUL_5037|M.ulcerans_Agy99 -------------MTPKRVGLYNPAFEHDSCGVAMVVDMHGRRSRDIVDK
MLBr_00061|M.leprae_Br4923 -------------MTAKRVGLYNPAFEHDSCGVAMVVDMYGRRTRDIVDK
Mflv_1114|M.gilvum_PYR-GCK -------------MAPSRQGLYDPAFEHDSCGVAMVADIHGRRSRDIVEK
Mvan_5698|M.vanbaalenii_PYR-1 ----------------------------------MVADMHGRRSRDIVDK
MSMEG_6459|M.smegmatis_MC2_155 MGMTPSSFGGSGRSSRDNRGLYNPAYEHDACGVAMVADMHGRRSRDIVDK
TH_2102|M.thermoresistible__bu -------------MTPKAQGLYNPAFEHDACGVAMVADMYGRRSRDIVDK
MAB_2486c|M.abscessus_ATCC_199 --------------------------------------------------
Mb3889c|M.bovis_AF2122/97 AITALLNLEHRGAQGAEPRSGDGAGILIQVPDEFLREAV----DFELPAP
Rv3859c|M.tuberculosis_H37Rv AITALLNLEHRGAQGAEPRSGDGAGILIQVPDEFLREAV----DFELPAP
MAV_0166|M.avium_104 AITALLNLEHRGAQGAEPRSGDGAGILIQVPDAFLREVV----DFELPPE
MMAR_5413|M.marinum_M AITALLNLEHRGAAGAEPRSGDGAGILIQVPDAFLREVV----DFELPAP
MUL_5037|M.ulcerans_Agy99 AITALLNLEHRGAAGAEPRSGDGAGILIQVPDAFLREVV----DFELPAP
MLBr_00061|M.leprae_Br4923 AITALLNLEHRGAQGAEPRSGDGAGILIQVPDAFLRAVV----DFELPAA
Mflv_1114|M.gilvum_PYR-GCK AITALLNLEHRGAQGAEPNTGDGAGILLQVPDEFFRAVC----GFDLPEP
Mvan_5698|M.vanbaalenii_PYR-1 AITALLNLEHRGAQGAEPNTGDGAGILLQVPDEFFRAVCEKE-GFDLPEA
MSMEG_6459|M.smegmatis_MC2_155 AITALVNLEHRGAQGAEPNTGDGAGILIQVPDAFLRAVV----DFDLPEP
TH_2102|M.thermoresistible__bu AITALLNLEHRGAQGAEPNTGDGAGILLQVPDEFLRAVVEEQGDFELPPP
MAB_2486c|M.abscessus_ATCC_199 --------------MSSEGVGDGAGVNLDLSLRFFSELT----GVSDLRH
:. *****: :::. *: ...
Mb3889c|M.bovis_AF2122/97 GSYATGIAFLPQSSKDAAAACAAVQKIAEAEGLQVLGWRSVPTDDSSLGA
Rv3859c|M.tuberculosis_H37Rv GSYATGIAFLPQSSKDAAAACAAVQKIAEAEGLQVLGWRSVPTDDSSLGA
MAV_0166|M.avium_104 GSYATGIAFLPQSSKDAATACAAVEKIAEAEGLTVLGWRNVPTDDSSLGA
MMAR_5413|M.marinum_M GSYATGIAFLPQSSKDAAAACAAVEKIAEAEGLQVLGWRNVPTDDSSLGA
MUL_5037|M.ulcerans_Agy99 GSYATGIAFLPQSSKDAAAACAAVEKIGEAEGLQVLGWRNVPTDDSSLGA
MLBr_00061|M.leprae_Br4923 SSYATGIAFLPQSSKDAAAACAAVEKIAVAEGLHVLGWRNVPTDASSLGA
Mflv_1114|M.gilvum_PYR-GCK GSFATGIAFLPQSAKDAATACESVEKIAEAEGLRVLGWRDVPTDDSSLGA
Mvan_5698|M.vanbaalenii_PYR-1 GSYATGMAFLPQSSKDAAIACAAVEKIAEAEGLQVLGWRDVPTDDSSLGA
MSMEG_6459|M.smegmatis_MC2_155 GSYATGMAFLPQSSRDAAAACESVEKIAEAEGLEVLGWREVPTDDSSLGA
TH_2102|M.thermoresistible__bu GSYATGIAFLPQSAKDAAQACDAVEKIVEAEGLTVLGWREVPIDDSSLGA
MAB_2486c|M.abscessus_ATCC_199 GQFGVGNFFLPNDPASHAEAEQVIEQALRAQGFTILLSRDVPVNNDAIRP
..:..* ***:.. . * * ::: *:*: :* *.** : .:: .
Mb3889c|M.bovis_AF2122/97 LSRDAMPTFRQVFLAGASGMALERRCYVVRKRAEHELGTKGPGQDGPGRE
Rv3859c|M.tuberculosis_H37Rv LSRDAMPTFRQVFLAGASGMALERRCYVVRKRAEHELGTKGPGQDGPGRE
MAV_0166|M.avium_104 LSRDAMPTFRQVFMAGASGMTLERRAYLVRKRAEHELGTKGPGQDGPGRE
MMAR_5413|M.marinum_M LSRDAMPTFRQVFLAGASDMTLERRCYVIRKRAEHELGTKGPGQDGPGRE
MUL_5037|M.ulcerans_Agy99 LSRDAMPTFRQVFLAGASDMALERRCYVIRKRAEHELGTKGPGQDGPGRE
MLBr_00061|M.leprae_Br4923 LSRDAMPTFRQVFMAGATGLTLERRAYVIRKRAEHELGTKGPGQDGPGRE
Mflv_1114|M.gilvum_PYR-GCK LARDAMPTFRQVFMAGAEGMDLERRAYVVRKRAEHELGTKGPGQDGPGRE
Mvan_5698|M.vanbaalenii_PYR-1 LARDAMPTFRQIFLAGASGMELERRAYVVRKRAEHELGTKGPGQDGPGRE
MSMEG_6459|M.smegmatis_MC2_155 LARDAMPTFRQLFLGGASGMELERRAYVVRKRAEHELGTKGPGQDGPGRE
TH_2102|M.thermoresistible__bu LARDAMPTFRQLFLGGATGIDLERRAYVVRKRAEHELGTKGPGQDGPGRE
MAB_2486c|M.abscessus_ATCC_199 AAIKYQLPIRQWVFRSDS----------ERKTHEALMAIESVAYTRPELL
: . .:** .: . ** * :. :. . *
Mb3889c|M.bovis_AF2122/97 TVYFPSLSGQTLVYKGMLTTPQLKAFYLDLQDERLTSALGIVHSRFSTNT
Rv3859c|M.tuberculosis_H37Rv TVYFPSLSGQTLVYKGMLTTPQLKAFYLDLQDERLTSALGIVHSRFSTNT
MAV_0166|M.avium_104 TVYFPSLSGQTFVYKGMLTTPQLKAFYLDLQDDRLTSALGIVHSRFSTNT
MMAR_5413|M.marinum_M TVYFPSLSGRTFVYKGMLTTPQLKAFYLDLQDDRLTSALGIVHSRFSTNT
MUL_5037|M.ulcerans_Agy99 TVYFPSLSGRTFVYKGMLTTPQLKAFYLDLQDDRLTSALGIVHSRFSTNT
MLBr_00061|M.leprae_Br4923 TIYFPSLSSQTFVYKGMLTTPQLKAFYRDLQDERLTSALGIVHSRFSTNT
Mflv_1114|M.gilvum_PYR-GCK TVYFPSLSGQTFVYKGMLTTPQLKAFYLDLQDERMTSALGIVHSRFSTNT
Mvan_5698|M.vanbaalenii_PYR-1 TVYFPSLSGQTFVYKGMLTTPQLKAFYLDLQDERMTSALGIVHSRFSTNT
MSMEG_6459|M.smegmatis_MC2_155 TVYFPSLSGQTFVYKGMLTTPQLKQFYLDLQDERLESALGIVHSRFSTNT
TH_2102|M.thermoresistible__bu TVYFPSLSGQTFVYKGMLTTPQLKAFYLDLQDERLTSALAIVHSRFSTNT
MAB_2486c|M.abscessus_ATCC_199 GLYPLSLSTRTQVLKGRLNSNEVVPYFSDLSDPRMEVHSMFFHTRFSTNT
:* *** :* * ** *.: :: :: **.* *: :.*:******
Mb3889c|M.bovis_AF2122/97 FPSWPLAHPFRRIAHNGEINTVTGNENWMRAREALIKTDIFGSAADVEKL
Rv3859c|M.tuberculosis_H37Rv FPSWPLAHPFRRIAHNGEINTVTGNENWMRAREALIKTDIFGSAADVEKL
MAV_0166|M.avium_104 FPSWPLAHPFRRIAHNGEINTVTGNENWMRAREALIKTDVFGTEADVEKL
MMAR_5413|M.marinum_M FPSWPLAHPFRRIAHNGEINTVTGNENWMRAREALIKTDVFGTEANVEKL
MUL_5037|M.ulcerans_Agy99 FPSWPLAHPFRRIAHNGEINTVTGNENWMRAREALIKTDVFGTEANVEKL
MLBr_00061|M.leprae_Br4923 FPSWPLAHPFRRIAHNGEINTVTGNENWMRAREALMKTDIFGSKADMEKL
Mflv_1114|M.gilvum_PYR-GCK FPSWPLAHPFRRVAHNGEINTVTGNENWMRAREALIKTDVFGAKADLDKI
Mvan_5698|M.vanbaalenii_PYR-1 FPSWPLAHPFRRVAHNGEINTVTGNENWMRAREALIKTDIFGGQ-DLEKV
MSMEG_6459|M.smegmatis_MC2_155 FPSWPLAHPFRRIAHNGEINTVTGNENWMRAREALIRTDVFGSD-GLDKI
TH_2102|M.thermoresistible__bu FPSWPLAHPFRRIAHNGEINTVTGNENWMRAREALIRTDLFGGQ-DLEKV
MAB_2486c|M.abscessus_ATCC_199 APNATMAQPFRLMAHNGELNTDKKNR---------LSENAIARARNRAII
*. .:*:*** :*****:** . *. : : :. . :
Mb3889c|M.bovis_AF2122/97 FPICTPGASDTARFDEVLELLHLGG-RSLAHAVLMMIPEAWERHESMDPA
Rv3859c|M.tuberculosis_H37Rv FPICTPGASDTARFDEVLELLHLGG-RSLAHAVLMMIPEAWERHESMDPA
MAV_0166|M.avium_104 FPICTPGASDTARFDEVLELLHLGG-RSLAHAVLMMIPEAWERHESMDPA
MMAR_5413|M.marinum_M FPICTPGASDTARFDEVLELLHLGG-RSLAHAVLMMIPEAWERHASMDPS
MUL_5037|M.ulcerans_Agy99 FPICTPGASDTARFDEVLELLHLGG-RSLAHAVLMMIPEAWERHASMDPS
MLBr_00061|M.leprae_Br4923 FPICTPGASDTARFDEALELLHLGG-RSLVHAVLMMIPEAWERHESMDPA
Mflv_1114|M.gilvum_PYR-GCK TPICTPGASDTARFDEVLELLHLGG-RSLPHAVLMMIPEAWERHESMDPA
Mvan_5698|M.vanbaalenii_PYR-1 TPICTPGASDTARFDEVLELLHLGG-RSLPHAVLMMIPEAWERHESMDPA
MSMEG_6459|M.smegmatis_MC2_155 VPVCTPGASDTARFDEVLELLHLGG-RSLPHAVLMMIPEAWERHEDMDPA
TH_2102|M.thermoresistible__bu APICTPGASDTARFDEVLELLHLGG-RSLPHAVLMMIPEAWEQHESMDPA
MAB_2486c|M.abscessus_ATCC_199 RPVGQ---SDSCRLDQTLQSRVLEDDLDLVTAVVAMMPPAWENDTTLSPR
*: **:.*:*:.*: * . .* **: *:* ***.. :.*
Mb3889c|M.bovis_AF2122/97 RRAFYQYHASLMEPWDGPASMTFTDGTVVGAVLDRNGLRPSRIWVTDDGL
Rv3859c|M.tuberculosis_H37Rv RRAFYQYHASLMEPWDGPASMTFTDGTVVGAVLDRNGLRPSRIWVTDDGL
MAV_0166|M.avium_104 RRAFYQYHASLMEPWDGPASMTFTDGTVIGAVLDRNGLRPSRIWVTEDGL
MMAR_5413|M.marinum_M RRAFYQYHSSLMEPWDGPASMTFTDGTIIGAVLDRNGLRPSRIWVTEDGL
MUL_5037|M.ulcerans_Agy99 RRAFYQYHSSLMEPWDGPASMTFTDGTMVGAVLDRNGLRPSRIWVTEDGL
MLBr_00061|M.leprae_Br4923 RRAFYQYHASLMEPWDGPASMTFTDGVVVGAVLDRNGLRPSRIWITDAGL
Mflv_1114|M.gilvum_PYR-GCK RRAFYEFHDSLMEPWDGPAAVCFTDGTVIGAVLDRNGLRPSRIWVTEDGL
Mvan_5698|M.vanbaalenii_PYR-1 RRAFYQYHASLMEPWDGPAAVCFTDGTVVGAVLDRNGLRPSRIWVTSDGL
MSMEG_6459|M.smegmatis_MC2_155 RRAFYEYHASLMEPWDGPASVCFTDGTVIGAVLDRNGLRPSRIWVTDDGL
TH_2102|M.thermoresistible__bu RRAFYEFHDSLMEPWDGPASVCFTDGTVIGAVLDRNGLRPSRIWVTADGL
MAB_2486c|M.abscessus_ATCC_199 VTAMLEYFSLYEEKNDGPAALIFGDGNIIGARLDRLGLRPLRTVETAEYL
*: ::. * ****:: * ** ::** *** **** * * *
Mb3889c|M.bovis_AF2122/97 VVMASEAGVLDLHPSTVVRRMRLQPGRMFLVDTAQGRIVSDEEIKADLAA
Rv3859c|M.tuberculosis_H37Rv VVMASEAGVLDLHPSTVVRRMRLQPGRMFLVDTAQGRIVSDEEIKADLAA
MAV_0166|M.avium_104 VVMASEAGVLDLDPATVVRRMRLQPGRMFLVDTAQGRIVSDEEIKAELAA
MMAR_5413|M.marinum_M VVMASEAGVLDLDPSTVVQRMRLQPGRMFLVDTAQGRIVSDEEIKAELAA
MUL_5037|M.ulcerans_Agy99 VVMASEAGVLDLDPSTVVQRMRLQPGRMFLVDTAQGRIVSDEAIKAELAA
MLBr_00061|M.leprae_Br4923 VVMASEAGVLDLDPSIVVRRMRLQPGRMFLVDTAQGRIVSDEEIKAKLAA
Mflv_1114|M.gilvum_PYR-GCK VVMASEAGVLDLDPSTVVKKMRLQPGRMFLVDTAQGRIVDDEEIKAQLAA
Mvan_5698|M.vanbaalenii_PYR-1 VVMASEAGVLDLDPGTVVQKMRLQPGRMFLVDTAQGRIVSDEEIKAELAA
MSMEG_6459|M.smegmatis_MC2_155 VVMASEAGVLPLDPAKVVKKMRLQPGRMFLVDTAQGRIVDDEEVKAELAA
TH_2102|M.thermoresistible__bu VVMASEAGVLNLDPATVVRKMRLQPGRMFLVDTAQGRIIDDEEIKAELAA
MAB_2486c|M.abscessus_ATCC_199 GVMS-EAGQIYFEPDSVLERGRIEAGGMRYYDHSEGRSYNTVEALEMLAA
**: *** : :.* *:.: *::.* * * ::** . ***
Mb3889c|M.bovis_AF2122/97 EHPYQEWLDNGLVPLDELPEGKDVRMPHHRIVM-------RQLAFGYTYE
Rv3859c|M.tuberculosis_H37Rv EHPYQEWLDNGLVPLDELPEGKDVRMPHHRIVM-------RQLAFGYTYE
MAV_0166|M.avium_104 EHPYQEWLDRNLVPLDDLPQGDYKRMPHDRLVK-------RQQTFGYTYE
MMAR_5413|M.marinum_M EHPYQEWLDNNLVPLEKLPSGEYARMAHERVVL-------RQLVFGYTYE
MUL_5037|M.ulcerans_Agy99 EHPYQEWLDNNLVPLEKLPSGEYARMAHERVVL-------RQLVFGYTYE
MLBr_00061|M.leprae_Br4923 EHPYQEWLDKGLVPIDELPAGKYVRMPHHRLVM-------RQLAFGYTYE
Mflv_1114|M.gilvum_PYR-GCK EHPYQEWLDAGLFQLDELPPGDYVRMPHHRVVL-------RQQIFGYTYE
Mvan_5698|M.vanbaalenii_PYR-1 GQPYQEWLDAGLFHLDELPQGDYVRMPHHRVVL-------RQQIFGYTYE
MSMEG_6459|M.smegmatis_MC2_155 AEPYQEWLEQGLFHLDELPPGDYVRMPHHRVVL-------RQQIFGYTYE
TH_2102|M.thermoresistible__bu EHPYQEWLDAGLFPLDELPQGDYQRMPHHRVVL-------RQRVFGYTSE
MAB_2486c|M.abscessus_ATCC_199 RHDYPAMLAQARVNIADLPHVPPAGQGSPARYDGDLDRHQRYVAYSLNQE
. * * . : .** * :. . *
Mb3889c|M.bovis_AF2122/97 ELNLLVAPMARLGAEPIGSMGTDTPVAVLSQRPRMLYDYFHQLFAQVTNP
Rv3859c|M.tuberculosis_H37Rv ELNLLVAPMARLGAEPIGSMGTDTPVAVLSQRPRMLYDYFHQLFAQVTNP
MAV_0166|M.avium_104 ELNLLVAPMVRTGAEPIGSMGTDTPVAVLSQRPRMLYDYFQQLFAQVTNP
MMAR_5413|M.marinum_M ELNLLVAPMVRTGAEPIGSMGTDTPIAVLSRRPRMLYDYFQQLFAQVTNP
MUL_5037|M.ulcerans_Agy99 ELNLLVAPMVRTGAEPIGSMGTDTPIAVLSRRPRMLYDYFQQLFAQVTNP
MLBr_00061|M.leprae_Br4923 ELNLLVAPMVRAGSEPIGSMGADTPIAVLSRRPRMLYDYFHQLFAQVTNP
Mflv_1114|M.gilvum_PYR-GCK ELNLLVAPMARNGAEALGSMGTDTPIAVLSSRPRMLYDYFQQLFAQVTNP
Mvan_5698|M.vanbaalenii_PYR-1 ELNLLVAPMARSGAEALGSMGTDTPIAVLSARPRMLFDYFQQLFAQVTNP
MSMEG_6459|M.smegmatis_MC2_155 ELNLLVAPMARTGAEALGSMGTDTPIAVLSARPRMLFDYFQQLFAQVTNP
TH_2102|M.thermoresistible__bu ELNLIVGPMARTGAEPLGSMGTDTPIAVLSQRSRLLFDYFQQMFAQVTNP
MAB_2486c|M.abscessus_ATCC_199 SFKFLMDPMLATGQEKISAMGYGAAINALSNQEGGVAKYFSQRFAQVTNP
.::::: ** * * :.:** .:.: .** : : .** * *******
Mb3889c|M.bovis_AF2122/97 PLDAIREEVVTSLQGTTGGERDLLNPDENSCHQIVLPQPILRNHELAKLV
Rv3859c|M.tuberculosis_H37Rv PLDAIREEVVTSLQGTTGGERDLLNPDQNSCHQIVLPQPILRNHELAKLV
MAV_0166|M.avium_104 PLDAIREEVVTSLQGTTGGERDLLTPTELSCHQIVLSQPILRNRELAKLV
MMAR_5413|M.marinum_M PLDAIREEVVTSLQGTTGGERDLLNPDENSCHQIQLPQPILRNHELAKLI
MUL_5037|M.ulcerans_Agy99 PLDAIREEVVTSLQGTTGGERDLLNPDENSCHQIQLPQPILRNHELAKLI
MLBr_00061|M.leprae_Br4923 PLDAIREEVVTSLQGTTGGEYDLLSPDENSCHQIWLPQPILRNYELAKLV
Mflv_1114|M.gilvum_PYR-GCK PLDAIREEVVTSLQGAMGPEGDLLNPGPESCRQIVLPQPILRNAELSKLI
Mvan_5698|M.vanbaalenii_PYR-1 PLDAIREEVVTSLQGTVGPEGDLLNPDAESCRQIVLPNPILRNAELSKLI
MSMEG_6459|M.smegmatis_MC2_155 PLDAIREEVVTSLQGTVGPEGDLLNPGPESCRQIVLPQPILRNADLSKLI
TH_2102|M.thermoresistible__bu PLDAIREEVITSLQGVIGPEGDLLNPGPESCRQIVLPQPILRNHELAKLI
MAB_2486c|M.abscessus_ATCC_199 PLDSIREADGMTLRVALGAKP---NSGAQPAPQIVVPSPILTHLDMLKIR
***:*** :*: . * : .. .. ** :..*** : :: *:
Mb3889c|M.bovis_AF2122/97 SLDPNDKVNGRPHGLRSKVIRCLYRVSEGGAGLAAALEEVRGAAAAAIAD
Rv3859c|M.tuberculosis_H37Rv SLDPNDKVNGRPHGLRSKVIRCLYRVSEGGAGLAAALEEVRGAAAAAIAD
MAV_0166|M.avium_104 NLNPDDEVNGRPHGMRSKVIRCLYPVAEGGAGLAAALEDVRAQASAAIAD
MMAR_5413|M.marinum_M NLDPETVINGRPHGLKSKVVRCLYPVAEGGAGLAAALNEVRAQASAAIAD
MUL_5037|M.ulcerans_Agy99 NLDPETVINGRPHGLKSKVVRCLYPVAEGGSGLAAALNEVRAQASAAIAD
MLBr_00061|M.leprae_Br4923 NLDPADEVNGRPHGMRSTVVSCLYPVADAGAGLAAALTEVRAQASAAIVG
Mflv_1114|M.gilvum_PYR-GCK CVDPDHEIRGHKHGMRAAVIRCLYPVNRGGQGLKEALDNVRAKVSSAIRD
Mvan_5698|M.vanbaalenii_PYR-1 CVDPDHEIRGHKHGMRAAVIRCLYPVNRGGRGLKEALDNVRAKVSAAIRD
MSMEG_6459|M.smegmatis_MC2_155 CVDPDHEIRGHKHGMRAAVIRCLYPVNRGGVGLKEALDNVRAKVSEAIRE
TH_2102|M.thermoresistible__bu NVDPDHEVRGRKHGMRAAVVRCLFPVNRGGQGLKEALDRVRAKVSAAIRE
MAB_2486c|M.abscessus_ATCC_199 EQG---RTPVRRFGMRYRVD--LTDAVSNAERIVRAIDDLCDAVEEFARE
. : .*:: * * . . : *: : .
Mb3889c|M.bovis_AF2122/97 GARIIILSDRESDEEMAPIPSLLAVAGVHHHLVRERTRTQVGLVVESGDA
Rv3859c|M.tuberculosis_H37Rv GARIIILSDRESDEEMAPIPSLLAVAGVHHHLVRERTRTQVGLVVESGDA
MAV_0166|M.avium_104 GARVIILSDRESDEKMAPIPSLLAVAGVHHHLVRDRTRTHVGLVVESGDA
MMAR_5413|M.marinum_M GARIIVLSDRESDERMAPIPALLAVAGVHHHLVRERSRTKVGLVVESGDA
MUL_5037|M.ulcerans_Agy99 GARIIVLSDRESDERMAPIPALLAVAGVHHHLVRERTRTKVGLVVESGDA
MLBr_00061|M.leprae_Br4923 GARVIILSDRNSDEQMAPIPSLLSVAAVHHHLVRDRTRTHVGLVVESGDA
Mflv_1114|M.gilvum_PYR-GCK GARIIVLSDRESDEQMAPIPSLLSVSAVHHHLVRERTRTQVGLIVEAGDA
Mvan_5698|M.vanbaalenii_PYR-1 GARIIVLSDRESNEQMAPIPSLLSVSAVHHHLVRDRTRTKVGLVVEAGDA
MSMEG_6459|M.smegmatis_MC2_155 GARIIVLSDRESNETMAPIPSLLSVAAVHHHLVRERTRTKVGLVVEAGDA
TH_2102|M.thermoresistible__bu GARIIVLSDRESDEQLAPIPSLLSVAAVHHHLVRDRTRTQVGLVVEAGDA
MAB_2486c|M.abscessus_ATCC_199 TGGIAVISDRHVSSERAAMPLIIVISAINQRLIEEGLRLRVSLIAESGQL
. : ::***. .. *.:* :: ::.::::*:.: * :*.*:.*:*:
Mb3889c|M.bovis_AF2122/97 REVHHMAALVGFGAAAINPYLVFESIEDMLDRGVIEGIDRTAALNNYIKA
Rv3859c|M.tuberculosis_H37Rv REVHHMAALVGFGAAAINPYLVFESIEDMLDRGVIEGIDRTAALNNYIKA
MAV_0166|M.avium_104 REVHHMAALVGFGAAAINPYMVFESIEDMLDRGVIEGIDRNTALNNYVKA
MMAR_5413|M.marinum_M REVHHMAMLLGCGAAAVNPYLAFESIEDMLDRGVIDGIDRNTALSNYVKA
MUL_5037|M.ulcerans_Agy99 REVHHMAMLLGCGAAAVNPYLAFASIEDMLDRGVIDGIDRNTALSNYVKA
MLBr_00061|M.leprae_Br4923 REVHHMAALIGFGAAAVNPYLVFESIEEMLDRGVIDGIDRKTALNNYIKA
Mflv_1114|M.gilvum_PYR-GCK REVHHMAALCGFGAAAINPYMAFESIEDMVDRGVITDITSDQAKANYVKA
Mvan_5698|M.vanbaalenii_PYR-1 REVHHMAALCGFGAAAINPYMAFESIEDMVDRGVISGISSDQAKANYVKA
MSMEG_6459|M.smegmatis_MC2_155 REVHHMATLVGFGAAAINPYMAFESIEDMVDRGVITGLTSDQAKANYVKA
TH_2102|M.thermoresistible__bu REVHHMAALIGFGASAINPYLAFESIEDMLDRNLLEGLDRETALNNYIKA
MAB_2486c|M.abscessus_ATCC_199 CSSHHVAAVLGFGASAVYPLAVQMRAEEKYGPDVDDAVN---AFKHFAKA
. **:* : * **:*: * . *: . .: : * :: **
Mb3889c|M.bovis_AF2122/97 AGKGVLKVMSKMGISTLASYTGAQLFQA--VGISEQVLDEYFTGLTCPTG
Rv3859c|M.tuberculosis_H37Rv AGKGVLKVMSKMGISTLASYTGAQLFQA--VGISEQVLDEYFTGLTCPTG
MAV_0166|M.avium_104 AGKGVLKVMSKMGISTLASYTGAQLFQA--VGISEDVLDEYFTGLSCPIG
MMAR_5413|M.marinum_M AGKGVLKVMSKMGISTLASYTGAQLFQA--VGVSEEVLNEYFSGLTCATG
MUL_5037|M.ulcerans_Agy99 AGKGVLKVMSKMGISTLASYTGAQLFQA--VGVSEEVLNEYFSGLTCATG
MLBr_00061|M.leprae_Br4923 TGQGVLKIMSKMGISTLASYVGAQLFQA--VGISEDVLDEYFSGLSCPTG
Mflv_1114|M.gilvum_PYR-GCK AGKGVLKVMSKMGISTLASYTGAQLFQA--IGINQQVLDDYFTGLTCPVG
Mvan_5698|M.vanbaalenii_PYR-1 AGKGVLKVMSKMGISTLASYTGAQLFQA--IGISQAVLDEYFAGLSCPVG
MSMEG_6459|M.smegmatis_MC2_155 AGKGVLKVMSKMGISTLASYTGAQLFQA--IGISQQVLDEYFTGLSCPVG
TH_2102|M.thermoresistible__bu AGKGVLKVMSKMGISTLRSYTGAQLFQA--IGISQEVLDEYFTGLSCPTG
MAB_2486c|M.abscessus_ATCC_199 AEKSLMKTMGRVGLCTVESYIGGEFFEPNYLDTSDPVLRKYFPNVVSPVA
: :.::* *.::*:.*: ** *.::*:. :. .: ** .**..: .. .
Mb3889c|M.bovis_AF2122/97 GITLDDIAADVAARHRLAYLDRPD--------------------------
Rv3859c|M.tuberculosis_H37Rv GITLDDIAADVAARHRLAYLDRPD--------------------------
MAV_0166|M.avium_104 GITLDDIAADVAARHALAYLDRPN--------------------------
MMAR_5413|M.marinum_M GITLDDIAADVATRHTLAYLDRPD--------------------------
MUL_5037|M.ulcerans_Agy99 GITLDDIAADVASRHTLAYLDRPD--------------------------
MLBr_00061|M.leprae_Br4923 GITLDDIAAEVAVRHRVAYLDRPD--------------------------
Mflv_1114|M.gilvum_PYR-GCK GIDLDDIAADIASRHALAYLDRPD--------------------------
Mvan_5698|M.vanbaalenii_PYR-1 GIDLDDIADDVATRHALAYLDRPD--------------------------
MSMEG_6459|M.smegmatis_MC2_155 GIDLEDIAADVAARHSLAFLDRPD--------------------------
TH_2102|M.thermoresistible__bu GIDLDDIAADVARRHELAYLDRPE--------------------------
MAB_2486c|M.abscessus_ATCC_199 GVGFSTLAAAVADWHARALAVTGEKDVPLLGLFKERAEGAGHSYGTTAVR
*: :. :* :* * * :
Mb3889c|M.bovis_AF2122/97 -------------ERAHRELEVGGEYQWRREGEYHLFN------------
Rv3859c|M.tuberculosis_H37Rv -------------ERAHRELEVGGEYQWRREGEYHLFN------------
MAV_0166|M.avium_104 -------------ERAHRELEVGGEYQWRREGEYHLFN------------
MMAR_5413|M.marinum_M -------------ERAHRELEVGGEYQWRREGEYHLFN------------
MUL_5037|M.ulcerans_Agy99 -------------ERAHRELEVGGEYQWRREGEYHLFN------------
MLBr_00061|M.leprae_Br4923 -------------ERAHRELEVGGEYQWRREGEYHLFN------------
Mflv_1114|M.gilvum_PYR-GCK -------------EWAHRELEVGGEYQWRREGEYHLFN------------
Mvan_5698|M.vanbaalenii_PYR-1 -------------EWAHRELEVGGEYQWRREGEYHLFN------------
MSMEG_6459|M.smegmatis_MC2_155 -------------ERAHRELEVGGEYQWRREGEYHLFN------------
TH_2102|M.thermoresistible__bu -------------EWAHRELEVGGEYQWRREGEYHLFN------------
MAB_2486c|M.abscessus_ATCC_199 GFVDMTEESISFGDEARPENPDMADPTYLRLLPLHQLEGAFGLDDEAYRN
: *: * .: : * * ::
Mb3889c|M.bovis_AF2122/97 -------------------PETVFKLQHSTRTGQYKIFKEYTRLVDDQ--
Rv3859c|M.tuberculosis_H37Rv -------------------PETVFKLQHSTRTGQYKIFKEYTRLVDDQ--
MAV_0166|M.avium_104 -------------------PETVFKLQHATRTGQYKIFKDYTRLVDDQ--
MMAR_5413|M.marinum_M -------------------PETVFKLQHSTRTGQYKIFKEYTRLVDDQ--
MUL_5037|M.ulcerans_Agy99 -------------------PETVFKLQHSTRTGQYKIFKEYTRLVDDQ--
MLBr_00061|M.leprae_Br4923 -------------------PETVFKLQHATRSGQYKIFKEYTRLVDDQ--
Mflv_1114|M.gilvum_PYR-GCK -------------------PDTVFKLQHSTRTGQYSIFKEYTQLVDDQ--
Mvan_5698|M.vanbaalenii_PYR-1 -------------------PDTVFKLQHSTRTGQYSVFKEYTSLVDDQ--
MSMEG_6459|M.smegmatis_MC2_155 -------------------PDTVFKLQHSTRTGQYSIFKEYTKLVDDQ--
TH_2102|M.thermoresistible__bu -------------------PDTVFKLQHSTRTGQYEIFKEYTRLVDDQ--
MAB_2486c|M.abscessus_ATCC_199 NSFDELPTRAIDSFDITPGYRNFVRAMNSERTRRPAALRDVLALPADVNF
...: :: *: : ::: * *
Mb3889c|M.bovis_AF2122/97 -------------------------------------------SERMASL
Rv3859c|M.tuberculosis_H37Rv -------------------------------------------SERMASL
MAV_0166|M.avium_104 -------------------------------------------SERMASL
MMAR_5413|M.marinum_M -------------------------------------------SERMASL
MUL_5037|M.ulcerans_Agy99 -------------------------------------------SERMASL
MLBr_00061|M.leprae_Br4923 -------------------------------------------SERMASL
Mflv_1114|M.gilvum_PYR-GCK -------------------------------------------SERMASL
Mvan_5698|M.vanbaalenii_PYR-1 -------------------------------------------SERMASL
MSMEG_6459|M.smegmatis_MC2_155 -------------------------------------------SERIASL
TH_2102|M.thermoresistible__bu -------------------------------------------SERMASL
MAB_2486c|M.abscessus_ATCC_199 VRTADEFRREMGQFARLGNNDFQVRGLSCQQTDDVFVLRLAHSPERLAAL
.**:*:*
Mb3889c|M.bovis_AF2122/97 RGLLKFRTG------------------------------VRPPVPLDEVE
Rv3859c|M.tuberculosis_H37Rv RGLLKFRTG------------------------------VRPPVPLDEVE
MAV_0166|M.avium_104 RGLLKFRAG------------------------------VRPPVPLEEVE
MMAR_5413|M.marinum_M RGLLKFRAG------------------------------VRPPIPLDEVE
MUL_5037|M.ulcerans_Agy99 RGLLKFRAG------------------------------VRPPIPLDEVE
MLBr_00061|M.leprae_Br4923 RGLLKFRSG------------------------------VRTPVPLDEVE
Mflv_1114|M.gilvum_PYR-GCK RGLLKFRDG------------------------------ERPSVPLDEVE
Mvan_5698|M.vanbaalenii_PYR-1 RGLLKFREG------------------------------VRPPVPLEEVE
MSMEG_6459|M.smegmatis_MC2_155 RGLLKFREG------------------------------HRPPVPIEEVE
TH_2102|M.thermoresistible__bu RGLLKFKEG------------------------------VRPPVPIEEVE
MAB_2486c|M.abscessus_ATCC_199 ASSLRTRFGREITHHEIVGDELRITATGQAADYLAQIRTAPESIPLSSVQ
. *: : * .:*:..*:
Mb3889c|M.bovis_AF2122/97 PASEIVKRFSTGAMSYGSISAEAHETLAIAMNRLGARSNCGEGGEDVKRF
Rv3859c|M.tuberculosis_H37Rv PASEIVKRFSTGAMSYGSISAEAHETLAIAMNRLGARSNCGEGGEDVKRF
MAV_0166|M.avium_104 PASEIVKRFSTGAMSYGSISAEAHETLAIAMNRLGGRSNSGEGGEDVKRF
MMAR_5413|M.marinum_M PASEIVKRFSTGAMSFGSISAEAHETLAIAMNRLGGRSNSGEGGEDVGRF
MUL_5037|M.ulcerans_Agy99 PASEIVKRFSTGAMSFGSISAEAHETLAIAMNRLGGRSNSGEGGEDVGRF
MLBr_00061|M.leprae_Br4923 PASEIVKRFSTGAMSYGSISAEAHETLAIAMNRLGGRSNSGEGGEDVNRF
Mflv_1114|M.gilvum_PYR-GCK PASEIVKRFSTGAMSYGSISAEAHETLAIAMNRLGGRSNSGEGGESVSRF
Mvan_5698|M.vanbaalenii_PYR-1 PASEIVKRFSTGAMSYGSISAEAHETLAIAMNRLGGRSNSGEGGESVTRF
MSMEG_6459|M.smegmatis_MC2_155 PASEIVKRFSTGAMSYGSISAEAHETLAIAMNRLGGRSNSGEGGEAVHRF
TH_2102|M.thermoresistible__bu PASEIVKRFSTGAMSYGSISAEAHETLAIAMNRLGARSNSGEGGEHVSRF
MAB_2486c|M.abscessus_ATCC_199 PASEITPTLTSGAMSHGALVAPAHEAVAHGTNLVGGLSNCGEGGEHISRY
*****. :::****.*:: * ***::* . * :*. **.***** : *:
Mb3889c|M.bovis_AF2122/97 DRDPNGDWRRSAIKQVASARFGVTSHYLTN--CTDLQIKMAQGAKPGEGG
Rv3859c|M.tuberculosis_H37Rv DRDPNGDWRRSAIKQVASARFGVTSHYLTN--CTDLQIKMAQGAKPGEGG
MAV_0166|M.avium_104 DRDPDGSWRRSAIKQVASGRFGVTSHYLTN--CTDIQIKMAQGAKPGEGG
MMAR_5413|M.marinum_M EQDPNGDWRRSAIKQVASARFGVTSHYLTN--CTDIQIKMAQGAKPGEGG
MUL_5037|M.ulcerans_Agy99 EQDPNGDWRRSAIKQVASARFGVTSHYLTN--CTDIQIKMAQGAKPGEGG
MLBr_00061|M.leprae_Br4923 EPDPNGDWRRSAIKQVASARFGVTSHYLTN--CTDIQIKMAQGAKPGEGG
Mflv_1114|M.gilvum_PYR-GCK DRDENGDWRRSAIKQVASGRFGVTSHYLTN--CTDIQIKMAQGAKPGEGG
Mvan_5698|M.vanbaalenii_PYR-1 DPDENGDWRRSAIKQVASGRFGVTSHYLAN--CTDIQIKMAQGAKPGEGG
MSMEG_6459|M.smegmatis_MC2_155 NPDENGDWRRSAIKQVASGRFGVTSHYLAN--CTDIQIKMAQGAKPGEGG
TH_2102|M.thermoresistible__bu DPDDNGDWRRSAIKQVASGRFGVTSHYLTN--CTDIQIKMAQGAKPGEGG
MAB_2486c|M.abscessus_ATCC_199 G-----TIRGSRIKQFASGRFGVWAGYLADPMLQELEIKIGQGAKPGEGG
* * ***.**.**** : **:: :::**:.*********
Mb3889c|M.bovis_AF2122/97 QLPGHKVYPWVAEVRHSTPGVGLISPPPHHDIYSIEDLAQLIHDLKNANP
Rv3859c|M.tuberculosis_H37Rv QLPGHKVYPWVAEVRHSTPGVGLISPPPHHDIYSIEDLAQLIHDLKNANP
MAV_0166|M.avium_104 QLPGHKVYPWVAEVRHSTPGVGLISPPPHHDIYSIEDLAQLIHDLKNANP
MMAR_5413|M.marinum_M QLPGHKVYPWVAKVRHSTPGVGLISPPPHHDIYSIEDLAQLIHDLKNANP
MUL_5037|M.ulcerans_Agy99 QLPGHKVYPWVAKVRHSTPGVGLISPPPHHDIYSIEDLAQLIHDLKNANP
MLBr_00061|M.leprae_Br4923 QLPGRKVYPWIAQVRNSTPGVGLISPPPHHDIYSIEDLAQLIHDLKNANP
Mflv_1114|M.gilvum_PYR-GCK QLPGHKVYPWVAEVRHSTPGVGLISPPPHHDIYSIEDLAQLIHDLKNANP
Mvan_5698|M.vanbaalenii_PYR-1 QLPGHKVYPWVAEVRHSTPGVGLISPPPHHDIYSIEDLAQLIHDLKNANP
MSMEG_6459|M.smegmatis_MC2_155 QLPGHKVYPWVAEVRHSTPGVGLISPPPHHDIYSIEDLAQLIHDLKNANP
TH_2102|M.thermoresistible__bu QLPGNKVYPWVAEVRHSTPGVGLISPPPHHDIYSIEDLAQLIHDLKNANP
MAB_2486c|M.abscessus_ATCC_199 QLPAPKVTVQIAAARGGTPGVELVSPPPHHDTYSIEDLAQLIHDCKAAR-
***. ** :* .* .**** *:******* ************ * *.
Mb3889c|M.bovis_AF2122/97 SARVHVKLVSENGVGTVAAGVSKAHADVVLISGHDGGTGATPLTSMKHAG
Rv3859c|M.tuberculosis_H37Rv SARVHVKLVSENGVGTVAAGVSKAHADVVLISGHDGGTGATPLTSMKHAG
MAV_0166|M.avium_104 AARVHVKLVSENGVGTVAAGVSKAHADVVLISGHDGGTGATPLTSMKHAG
MMAR_5413|M.marinum_M SARVHVKLVSENGVGTVAAGVSKAHADVVLISGHDGGTGATPLTSMKHAG
MUL_5037|M.ulcerans_Agy99 SARVHVKLVSENGVGTVAAGVSKAHADVVLISGHDGGTGATPLTSMKHAG
MLBr_00061|M.leprae_Br4923 SARVHVKLVSENGVGTVAAGVSKAHADVVLVSGHDGGTGATPLTSMKHSG
Mflv_1114|M.gilvum_PYR-GCK QARVHVKLVSENGVGTVAAGVSKAHADVVLISGHDGGTGATPLTSMKHAG
Mvan_5698|M.vanbaalenii_PYR-1 QARIHVKLVSENGVGTVAAGVSKAHADVVLISGHDGGTGATPLTSMKHAG
MSMEG_6459|M.smegmatis_MC2_155 AARIHVKLVSENGVGTVAAGVSKAHADVVLISGHDGGTGASPLTSLKHAG
TH_2102|M.thermoresistible__bu QARVHVKLVSEAGVGTVAAGVSKAHADVVLISGHDGGTGASPLTSLKHAG
MAB_2486c|M.abscessus_ATCC_199 -VRVIVKLVSSEGIGTIAVGVAKAGADVINVAGNTGGTGAAAVTSLKYAG
.*: *****. *:**:*.**:** ***: ::*: *****:.:**:*::*
Mb3889c|M.bovis_AF2122/97 APWELGLAETQQTLLLNGLRDRIVVQVDGQLKTGRDVMIATLLGAEEFGF
Rv3859c|M.tuberculosis_H37Rv APWELGLAETQQTLLLNGLRDRIVVQVDGQLKTGRDVMIATLLGAEEFGF
MAV_0166|M.avium_104 APWELGLAETQQTLLLNGLRDRIVVQVDGQLKTGRDVMIAALLGAEEFGF
MMAR_5413|M.marinum_M APWELGLAETQQTLLLNGLRDRIVVQVDGQLKTGRDVMIATLLGGEEFGF
MUL_5037|M.ulcerans_Agy99 APWELGLAETQQTLLLNGLRDRIVVQVDGQLKTGRDVMIATLLGGEEFGF
MLBr_00061|M.leprae_Br4923 APWELGLAETQQTLLLNGLRDRIVVQVDGQLKTGRDVMIAALLGAEEFGF
Mflv_1114|M.gilvum_PYR-GCK APWELGLAETQQTLLLNGLRDRIVVQVDGQLKTGRDVVVAALLGAEEFGF
Mvan_5698|M.vanbaalenii_PYR-1 APWELGLAETQQTLLLNGLRDRIVVQVDGQLKTGRDVVVAALLGAEEFGF
MSMEG_6459|M.smegmatis_MC2_155 APWELGLAETQQTLLLNGLRDRIVVQVDGQLKTGRDVVIAALLGAEEFGF
TH_2102|M.thermoresistible__bu APWELGLAETQQTLLLNGLRDRIVVQVDGQLKTGRDVVIAALLGAEEFGF
MAB_2486c|M.abscessus_ATCC_199 RSAEIGVAEVHQALCANGIRQKVVLRCSGAHQTASDVVKSALLGADSFEF
. *:*:**.:*:* **:*:::*:: .* :*. **: ::***.:.* *
Mb3889c|M.bovis_AF2122/97 ATAPLVVAGCIMMRVCHLDTCPVGVATQNPLLRERFTGKPEFVENFFMFI
Rv3859c|M.tuberculosis_H37Rv ATAPLVVAGCIMMRVCHLDTCPVGVATQNPLLRERFTGKPEFVENFFMFI
MAV_0166|M.avium_104 ATAPLVVSGCIMMRVCHLDTCPVGVATQNPVLRERFTGKPEFVENFFMFI
MMAR_5413|M.marinum_M ATAPLVVAGCIMMRVCHLDTCPVGVATQNPLLRERFNGKPEFVENFFMFI
MUL_5037|M.ulcerans_Agy99 ATAPLVVAGCIMMRVCHLDTCPVGVATQNPLLRERFNGKPEFVENFFMFI
MLBr_00061|M.leprae_Br4923 ATAPLVVAGCIMMRVCHLDTCPVGVATQNPLLRKRFNGKPEFVENFFMFI
Mflv_1114|M.gilvum_PYR-GCK ATAPLVVAGCIMMRVCHLDTCPVGVATQNPVLRKRFNGQPEFVENFFMFI
Mvan_5698|M.vanbaalenii_PYR-1 ATAPLVVTGCIMMRVCHLDTCPVGVATQNPLLRQRFNGKPEFVENFFMFI
MSMEG_6459|M.smegmatis_MC2_155 ATAPLVVSGCIMMRVCHLDTCPVGVATQNPLLRQRFNGKPEFVENFFMFI
TH_2102|M.thermoresistible__bu ATAPLVVSGCVMMRVCHLDTCPVGVATQNPVLRQRFTGKPEFVENFFLFI
MAB_2486c|M.abscessus_ATCC_199 GTTALMMLKCVMAKNCNIK-CPAGLTTN----AEVFEGDPRALAQYLLNI
.*:.*:: *:* : *::. **.*::*: : * *.*. : :::: *
Mb3889c|M.bovis_AF2122/97 AEEVREYLAQLGFRTVNEAVGQAGALDTTLARAHWKAHKLDLAPVLHEPE
Rv3859c|M.tuberculosis_H37Rv AEEVREYLAQLGFRTVNEAVGQAGALDTTLARAHWKAHKLDLAPVLHEPE
MAV_0166|M.avium_104 AEEVREYMAQLGFRTLNEAVGQVKSLDTTLARAHWKAHRLDLGPVLHEPE
MMAR_5413|M.marinum_M AEEVREYMAQLGFRTINEAVGQAGALDTTLARAHWKAHRLDLTPVLHEPE
MUL_5037|M.ulcerans_Agy99 AEEVREYMAQLGFRTINEAVGQAGALDTTLARAHWKAHRLDLTPVLHEPE
MLBr_00061|M.leprae_Br4923 AEEVREYMAQLGFRTFNEAVGQVGALDTTPVREHWKAHSIDLTPVLHEPE
Mflv_1114|M.gilvum_PYR-GCK AEEVRELMAELGFRTVNEMVGQVGALDTTQAAERWKAHKLDLTPVLHEPD
Mvan_5698|M.vanbaalenii_PYR-1 AEEVRELMAQLGFRTVNEMVGQVGALDTTQAAEHWKAHKLDLAPVLHEPE
MSMEG_6459|M.smegmatis_MC2_155 AEEVRELMAQLGFRTVNEMVGQVGALDTTKAAEHWKAHKLDLTPVLHEPD
TH_2102|M.thermoresistible__bu AEEVREYMAKMGFRTVNEMVGQVGMLDTTRAAEHWKAHKLDLSPVLHQPE
MAB_2486c|M.abscessus_ATCC_199 AHEVREILAALGMRTLREARGRSDLLHLLDHPSSIGTLDLRRMLAVAEEF
*.**** :* :*:**..* *: *. : : .: :
Mb3889c|M.bovis_AF2122/97 SAFMNQDLYCSSRQDHGLDKALDQQLIVMSREALDSGKPVRFSTTIGNVN
Rv3859c|M.tuberculosis_H37Rv SAFMNQDLYCSSRQDHGLDKALDQQLIVMSREALDSGKPVRFSTTIGNVN
MAV_0166|M.avium_104 SAFMNQDLYCSSRQDHGLDKALDQQLIVMSREALDSGKPVRFSTTISNVN
MMAR_5413|M.marinum_M SAFMNQDLYCSSSQDHGLDKALDQQLIVMSREALDSGKPVRFATTISNVN
MUL_5037|M.ulcerans_Agy99 SAFMNQGLYCSSSQDHGLDKALDQQLIVMSREALDSGKPVRFATTISNVN
MLBr_00061|M.leprae_Br4923 SAFMNQDLYCSSRQDHGLDKVLDQQLIAVSRDALDFGKPVRFSTTIGNVN
Mflv_1114|M.gilvum_PYR-GCK SAFMNQDLYCSSRQDHGLDKALDQQLITQSREAIDNGTPVRFSTTIANVN
Mvan_5698|M.vanbaalenii_PYR-1 SAFMNQDLYCSSRQDHGLDKALDQQLIAQCREALDRGTPVRFSTTIANVN
MSMEG_6459|M.smegmatis_MC2_155 SAFMNQDLYCSSRQDHGLDKALDQQLIVQSREALDSGTPVRFTAKITNVN
TH_2102|M.thermoresistible__bu SAFMNQDLYCSSRQDHGLDKALDQQLITMCREALDNGTPVRFSTTIANVN
MAB_2486c|M.abscessus_ATCC_199 VVENPVYMEKDYSVDDAFAAQFDARGAVLKPVTLTNQNKSVGGQLAIDIE
. : . *..: :* : . :: . :::
Mb3889c|M.bovis_AF2122/97 RTVGTMLGHELTKAYGGQG---LPDGTIDITFDGSAGNSFGAFVPKGITL
Rv3859c|M.tuberculosis_H37Rv RTVGTMLGHELTKAYGGQG---LPDGTIDITFDGSAGNSFGAFVPKGITL
MAV_0166|M.avium_104 RTVGTMLGHEVTKAYGGQG---LPDGTIDITFDGSAGNSFGAFVPRGITL
MMAR_5413|M.marinum_M RTVGTMLGHEVTKAYGGQG---LPDGSIDITFEGSAGNSFGAFVPKGITL
MUL_5037|M.ulcerans_Agy99 RTVGTMLGHEVTKAYGGQG---LPDGSIDITFEGSAGNSFGAFVPKGITL
MLBr_00061|M.leprae_Br4923 RTVGTMLGHELTKAYGAQG---LPDGTIDITFEGSAGNSFGAFVPKGITL
Mflv_1114|M.gilvum_PYR-GCK RTVGTMLGHEVTKAYGGQG---LPDGTIDITFEGSAGNSFGAFLPKGITL
Mvan_5698|M.vanbaalenii_PYR-1 RTVGTMLGHEVTKAYGGQG---LPDGTIDITFEGSAGNSFGAFVPKGITL
MSMEG_6459|M.smegmatis_MC2_155 RTVGTMLGHEVTKAYGGQG---LPDGTIDITFEGSAGNSFGAFVPKGITL
TH_2102|M.thermoresistible__bu RTVGTMLGHEVTKAYGGQG---LPDGTIDITFDGSAGNSFGAFVPRGITL
MAB_2486c|M.abscessus_ATCC_199 RSLNYQNIEGPAVATDERGRRYLLPESIAITTTGSAGQSYGVFCNDGMVL
*::. . : * . :* * :* ** ****:*:*.* *:.*
Mb3889c|M.bovis_AF2122/97 RVYGDANDYVGKGLSGGRIVVRPSDDAPQDYVAEDNIIGGNVILFGATSG
Rv3859c|M.tuberculosis_H37Rv RVYGDANDYVGKGLSGGRIVVRPSDDAPQDYVAEDNIIGGNVILFGATSG
MAV_0166|M.avium_104 RVYGDANDYVGKGLSGGRIVVRPSDNAPADYVAEDNIIGGNVILFGATSG
MMAR_5413|M.marinum_M RIYGDANDYVGKGLSGGRIVVRPSDNAPPGYVAEDNIIGGNVILFGATSG
MUL_5037|M.ulcerans_Agy99 RIYGDANDYVGKGLSGGRIVVRPSDNAPPGYVAEDNIIGGNVILFGATSG
MLBr_00061|M.leprae_Br4923 RVYGDANDYVGKGLSGGRIVVRPSDSVPLNYVAEDNIIGGNVILFGATSG
Mflv_1114|M.gilvum_PYR-GCK RVYGDANDYVGKGLSGGRVVVRPSDNAPEGYVAEDNIIAGNVVLFGATSG
Mvan_5698|M.vanbaalenii_PYR-1 RVYGDANDYVGKGLSGGRVVVRPSDGAPEGYAAEGNIIAGNVVLFGATSG
MSMEG_6459|M.smegmatis_MC2_155 RVYGDANDYVGKGLSGGRIVVRPADEAPESFAAEDNIIAGNVVLFGATSG
TH_2102|M.thermoresistible__bu RVYGDANDYVGKGLSGGRIVVRPPDNVPESFVAEDNIIAGNVILFGATSG
MAB_2486c|M.abscessus_ATCC_199 THTGTCNDGVGKSACGGTITVRSPGGGSS--EPGGNVLIGNFALFGASGG
* .** ***. .** :.**... . . .*:: **. ****:.*
Mb3889c|M.bovis_AF2122/97 EVYLRGVVGERFAVRNSGAHAVVEGVGDHGCEYMTGGRVVILGRTGRNFA
Rv3859c|M.tuberculosis_H37Rv EVYLRGVVGERFAVRNSGAHAVVEGVGDHGCEYMTGGRVVILGRTGRNFA
MAV_0166|M.avium_104 EAYLRGVVGERFAVRNSGAHAVVEGVGDHGCEYMTGGKVVVLGRTGRNFA
MMAR_5413|M.marinum_M EVFLRGVVGERFAVRNSGAHAVVEGVGDHGCEYMTGGRVVILGQTGRNFA
MUL_5037|M.ulcerans_Agy99 EVFLRGVVGERFAVRNSGAHVVVEGVGDHGCEYMTGGRVVILGQTGRNFA
MLBr_00061|M.leprae_Br4923 EVYLRGVVGERFAVRNSGAHAVVEGVGDHGCEYMTGGKVVILGRTGRNFA
Mflv_1114|M.gilvum_PYR-GCK EMFLRGQVGERFAVRNSGAHAVVEGVGDHGCEYMTGGRVVILGPTGRNFA
Mvan_5698|M.vanbaalenii_PYR-1 QMFLRGQVGERFAVRNSGAHAVVEGVGDHGCEYMTGGRVVILGPTGRNFA
MSMEG_6459|M.smegmatis_MC2_155 EAFLRGQVGERFAVRNSGAVAVVEGVGDHGCEYMTGGRVVILGPTGRNFA
TH_2102|M.thermoresistible__bu EVFLRGQVGERFAVRNSGALAVVEGVGDHGCEYMTGGRVVILGPVGRNFA
MAB_2486c|M.abscessus_ATCC_199 RLFVQGQAGDRFAVRNSGATAVVEGTGEFLCEYMTNGAVLNIGDFGKGVA
. :::* .*:********* .****.*:. *****.* *: :* *:..*
Mb3889c|M.bovis_AF2122/97 AGMSGGVAYVYDPDGELPANLNSEMVELETLDEDDA---DWLHGTIQVHV
Rv3859c|M.tuberculosis_H37Rv AGMSGGVAYVYDPDGELPANLNSEMVELETLDEDDA---DWLHGTIQVHV
MAV_0166|M.avium_104 AGMSGGVAYIYDPPGEFPQHLNAEMVELESLDDDDI---EWLHGMIQAHV
MMAR_5413|M.marinum_M AGMSGGIAYVYDPDGELPDNLNTEMVDVETLDDDDS---EFVHGIVQAHV
MUL_5037|M.ulcerans_Agy99 AGMSGGIAYVYDPDGELPDNLNTEMVDVETLDDDDS---EFVHGIVQAHV
MLBr_00061|M.leprae_Br4923 AGMSGGEAYVYDSDGELPANLNFEMVELEPLDFDDA---AWLYATIQAHV
Mflv_1114|M.gilvum_PYR-GCK AGMSGGVGYVYDPQDALAGNLNAEMVELESLGDSDSDDAVFVHGMIQAHV
Mvan_5698|M.vanbaalenii_PYR-1 AGMSGGIAYVYDPDNTLADNLNAEMVELEGMGGIDSDDAVFVHGIVQEHV
MSMEG_6459|M.smegmatis_MC2_155 AGMSGGIAYVYDPRAALPKNLNPEMVEIEALEEADT---EWLRGILASHV
TH_2102|M.thermoresistible__bu AGMSGGIAYVYDPDGVLPGRLNREMVELESLDDEDR---EWLRGFIAAHV
MAB_2486c|M.abscessus_ATCC_199 NGMSGGFLYQYDPHGQLPSKVSHDSVLVLPITDAPFH-EAAAHILLQWHV
***** * **. :. .:. : * : : : **
Mb3889c|M.bovis_AF2122/97 DATDSAVGQRILSDWSGQQRHFVKVMPRDYKRVLQAIALAERDGVD---V
Rv3859c|M.tuberculosis_H37Rv DATDSAVGQRILSDWSGQQRHFVKVMPRDYKRVLQAIALAERDGVD---V
MAV_0166|M.avium_104 DATDSAVGQRILGDWAQQQRHFVKVMPRDYKRVLQAIAEAERDGGD---V
MMAR_5413|M.marinum_M DATDSAVGQRVLADWPQQQRHFVKVMPRDYKRVLQAIAEAERDGVD---I
MUL_5037|M.ulcerans_Agy99 DATDSAVGQRVLADWPQQQRHFVKVMPRDYKRVLQAIAEAERDGVD---I
MLBr_00061|M.leprae_Br4923 DATDSAVGQRILAGWSEQQRYFVKVMPKDYKRVLQAIALAERNGVD---V
Mflv_1114|M.gilvum_PYR-GCK GATDSAVGQRILADWSGEQVHFKKVMPRDYKRVLEAIAEAERAGADENGV
Mvan_5698|M.vanbaalenii_PYR-1 DATDSAVGQRILADWNTELGHFKKVMPRDFKRVLQAIAEAQQSGADENGV
MSMEG_6459|M.smegmatis_MC2_155 DATDSAVGQRILADWDNELKHFAKVMPRDYKRVLAAIAEAEAKGLTGSDI
TH_2102|M.thermoresistible__bu DATDSAIGQRILNDWDEHVTHFAKVMPRDYKRVLTAIAEAEASGAD---V
MAB_2486c|M.abscessus_ATCC_199 AATGSTKGQALLDDWQSARDHMVYTMSRALLQYQDSDAILQGKTRKELLD
**.*: ** :* .* :: .*.: : : * :
Mb3889c|M.bovis_AF2122/97 DKAIMAAAHG----------------------------------------
Rv3859c|M.tuberculosis_H37Rv DKAIMAAAHG----------------------------------------
MAV_0166|M.avium_104 DKAIMAAAHG----------------------------------------
MMAR_5413|M.marinum_M DKAIMAASHG----------------------------------------
MUL_5037|M.ulcerans_Agy99 DKAIMAASHG----------------------------------------
MLBr_00061|M.leprae_Br4923 NKVIMEPAHG----------------------------------------
Mflv_1114|M.gilvum_PYR-GCK AEAIMAAANG----------------------------------------
Mvan_5698|M.vanbaalenii_PYR-1 AEAIMAAANG----------------------------------------
MSMEG_6459|M.smegmatis_MC2_155 DDAIMAASNG----------------------------------------
TH_2102|M.thermoresistible__bu DEAIMAAANG----------------------------------------
MAB_2486c|M.abscessus_ATCC_199 ELTAALAGYQVHKFKLSYRDRRDVVGGTVPAYGDTDTEGMYALLNTYTVL
. ..
Mb3889c|M.bovis_AF2122/97 --------------------------------------------------
Rv3859c|M.tuberculosis_H37Rv --------------------------------------------------
MAV_0166|M.avium_104 --------------------------------------------------
MMAR_5413|M.marinum_M --------------------------------------------------
MUL_5037|M.ulcerans_Agy99 --------------------------------------------------
MLBr_00061|M.leprae_Br4923 --------------------------------------------------
Mflv_1114|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5698|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_6459|M.smegmatis_MC2_155 --------------------------------------------------
TH_2102|M.thermoresistible__bu --------------------------------------------------
MAB_2486c|M.abscessus_ATCC_199 NMAQQLALSRMPNVTDVTDPRIGKAVRNLVLTEDFFLIQKLQKYAREAID
Mb3889c|M.bovis_AF2122/97 --------------------------------------------------
Rv3859c|M.tuberculosis_H37Rv --------------------------------------------------
MAV_0166|M.avium_104 --------------------------------------------------
MMAR_5413|M.marinum_M --------------------------------------------------
MUL_5037|M.ulcerans_Agy99 --------------------------------------------------
MLBr_00061|M.leprae_Br4923 --------------------------------------------------
Mflv_1114|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5698|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_6459|M.smegmatis_MC2_155 --------------------------------------------------
TH_2102|M.thermoresistible__bu --------------------------------------------------
MAB_2486c|M.abscessus_ATCC_199 GYSDEDLAVLIADKRLTDYKDALSQRNVLSMDSPGTYGWILHQSAKNIDK
Mb3889c|M.bovis_AF2122/97 -----------------------------
Rv3859c|M.tuberculosis_H37Rv -----------------------------
MAV_0166|M.avium_104 -----------------------------
MMAR_5413|M.marinum_M -----------------------------
MUL_5037|M.ulcerans_Agy99 -----------------------------
MLBr_00061|M.leprae_Br4923 -----------------------------
Mflv_1114|M.gilvum_PYR-GCK -----------------------------
Mvan_5698|M.vanbaalenii_PYR-1 -----------------------------
MSMEG_6459|M.smegmatis_MC2_155 -----------------------------
TH_2102|M.thermoresistible__bu -----------------------------
MAB_2486c|M.abscessus_ATCC_199 IGRLPSFEELFAHRALPAVALSGPSLQTT