For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VSDGTSGERERHEFVTLESEPVYMGGILALRRDRVAMPGGGSATREVVEHYGAVAVAAVDEAGRVALVYQ YRHPLGHRLWELPAGLLDIAGEDSQRAAARELLEEAGIEAGTWRVLVDAAASPGFTDEVVRVFLATGLSH PGRGESHDEEADMTVHWVPLSEAVSMVMAGEVVNAIAVAGILATHIALGGHHTRPVDAPWPDRPTAFAER KAAT
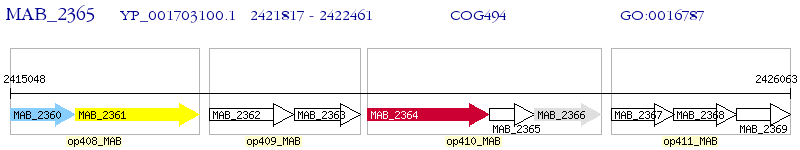
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_2365 | - | - | 100% (214) | MutT/nudix family protein |
| M. abscessus ATCC 19977 | MAB_4935 | - | 2e-07 | 29.41% (170) | MutT/NUDIX family protein |
| M. abscessus ATCC 19977 | MAB_3291 | - | 2e-05 | 30.43% (138) | MutT/NUDIX hydrolase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1726 | - | 3e-68 | 59.90% (202) | hypothetical protein Mb1726 |
| M. gilvum PYR-GCK | Mflv_3501 | - | 2e-67 | 63.82% (199) | NUDIX hydrolase |
| M. tuberculosis H37Rv | Rv1700 | - | 9e-69 | 59.90% (202) | hypothetical protein Rv1700 |
| M. leprae Br4923 | MLBr_02698 | - | 1e-05 | 28.17% (142) | hypothetical protein MLBr_02698 |
| M. marinum M | MMAR_2505 | - | 2e-67 | 62.19% (201) | NUDIX hydrolase |
| M. avium 104 | MAV_3071 | - | 4e-70 | 62.38% (202) | MutT/nudix family protein |
| M. smegmatis MC2 155 | MSMEG_3745 | - | 5e-69 | 65.50% (200) | MutT/nudix family protein |
| M. thermoresistible (build 8) | TH_1543 | - | 3e-51 | 62.26% (159) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_1689 | - | 1e-67 | 62.19% (201) | NUDIX hydrolase |
| M. vanbaalenii PYR-1 | Mvan_3282 | - | 1e-67 | 61.27% (204) | NUDIX hydrolase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1726|M.bovis_AF2122/97 ------------MAE-------------HDFETISSETLHTGAIFALRRD
Rv1700|M.tuberculosis_H37Rv ------------MAE-------------HDFETISSETLHTGAIFALRRD
MAV_3071|M.avium_104 ------------MAE-------------HVFETASSETLYTGKIFALRRD
MMAR_2505|M.marinum_M ------------MAE-------------HDFETISSETLYTGAIFALRRD
MUL_1689|M.ulcerans_Agy99 ------------MAE-------------HDFETISSETLYTGAIFALRRD
Mflv_3501|M.gilvum_PYR-GCK -------MAEDEPGL-------------HEFSTAASETVYVGKILALRGD
Mvan_3282|M.vanbaalenii_PYR-1 -------MAEGDPGR-------------HEFTTVASETVYAGKIIALRAD
MSMEG_3745|M.smegmatis_MC2_155 ------------MGE-------------HDFETLASETVYVGNIFALRAD
TH_1543|M.thermoresistible__bu --------------------------------------------------
MAB_2365|M.abscessus_ATCC_1997 -------MSDGTSGERER----------HEFVTLESEPVYMGGILALRRD
MLBr_02698|M.leprae_Br4923 MPAIIAWVSDGEQAKPRRRRKRRRGRDATRSAENHTDRQVANDATATKPR
Mb1726|M.bovis_AF2122/97 QVRMPGGGIVTREVVEHLGAVAIVAMDDNG----NIPMVYQYRHTYGRRL
Rv1700|M.tuberculosis_H37Rv QVRMPGGGIVTREVVEHFGAVAIVAMDDNG----NIPMVYQYRHTYGRRL
MAV_3071|M.avium_104 QVRMPGGKVVTREIVEHFGAVAVVAMDDDG----NIPMVYQYRHAFGRRL
MMAR_2505|M.marinum_M RVRMPGDTTAVREVIEHYGAVAIVAMDDNG----NIPMVYQYRHAFGRRL
MUL_1689|M.ulcerans_Agy99 RVRMPGDTTAVREVIEHYGAVAIVAMDDNG----NIPMVYQYRHAFGRRL
Mflv_3501|M.gilvum_PYR-GCK EVRMPGGGTARREVVEHFGAVAVVALDDEG----RVVLVHQYRHAFGRRL
Mvan_3282|M.vanbaalenii_PYR-1 EVRMPGGGTARREVIEHFGAVAVVALDEDG----RIVLIHQYRHAFGRRL
MSMEG_3745|M.smegmatis_MC2_155 EVSMPGGKSARREVVEHYGAVAVVALDDDG----NVVLVYQYRHPLGRRL
TH_1543|M.thermoresistible__bu -------------------------MDADR----NVALVHQYRHPVRRRL
MAB_2365|M.abscessus_ATCC_1997 RVAMPGGGSATREVVEHYGAVAVAAVDEAG----RVALVYQYRHPLGHRL
MLBr_02698|M.leprae_Br4923 RSHSPHRAPEQLRTVHETSAGGLVIYGIDGPRDAQVAALIGRIDRRGRML
. .: : . : *
Mb1726|M.bovis_AF2122/97 WELPAGLLDVAGEPPHLTAARELREEVGLQASTWQVLVDLDTAPGFSDES
Rv1700|M.tuberculosis_H37Rv WELPAGLLDVAGEPPHLTAARELREEVGLQASTWQVLVDLDTAPGFSDES
MAV_3071|M.avium_104 WELPAGLLDVHGEAAHLTAARELMEEAGLKAETWAVLVDLNSTPGFSDES
MMAR_2505|M.marinum_M LELPAGLRDAAGEPSHVTAARELHEEAGLQAEHWQVLIDLDSAPGFSDES
MUL_1689|M.ulcerans_Agy99 LELPAGLRDAAGEPSHVTAARELHEEAGLQAEHWQVLIDLDSAPGFSDES
Mflv_3501|M.gilvum_PYR-GCK WELPAGLLDSAGEDPHVTAARELVEEVGLAARDWWTLVDIDSAPGFCDES
Mvan_3282|M.vanbaalenii_PYR-1 WELPAGLLDFGGEPPHASAVRELAEEAGLAAEHWRTLIDVDSAPGFCDES
MSMEG_3745|M.smegmatis_MC2_155 WELPAGLLDLGGEPPEVTAARELEEEVGLAASDWRVLVDLDSTPGFSDES
TH_1543|M.thermoresistible__bu WELPAGLLDEPGEPPHLTAARELEEEAGLAARDWRVLVDLVSSPGFSDEC
MAB_2365|M.abscessus_ATCC_1997 WELPAGLLDIAGEDSQRAAARELLEEAGIEAGTWRVLVDAAASPGFTDEV
MLBr_02698|M.leprae_Br4923 WSLPKGHIE-QGETAEQTAIREVAEETGIRGSVLAALGQIDYWFVTDDCR
.** * : ** .. :* **: **.*: . .* : *
Mb1726|M.bovis_AF2122/97 VRVYLATGLREVGRPEAHHEEADMT-MGWYPIAEAARRVLRGEIVNSIAI
Rv1700|M.tuberculosis_H37Rv VRVYLATGLREVGRPEAHHEEADMT-MGWYPIAEAARRVLRGEIVNSIAI
MAV_3071|M.avium_104 VRVYLATGLTRVDRPEAHDEEADMT-LEWYPLADAARKVLSGEIVNAIAV
MMAR_2505|M.marinum_M VRVYLATGLSEVEQPEGHHEEADMT-VRWFPLTEAVSKVFTGEIVNAIAV
MUL_1689|M.ulcerans_Agy99 VRVYLATGLSEVEQPEGHHEEADMT-VRWFPLTEAVSKVFTGEIVNAIAV
Mflv_3501|M.gilvum_PYR-GCK VRIFLATGLSDVDRPEAHDEEADLA-VRRVDLAEAVARVFSGDIVNSIAV
Mvan_3282|M.vanbaalenii_PYR-1 VRVFLATGLRDVERPQAHDEEADMT-VQRFDLADAVAKVYSGEIVNSSAV
MSMEG_3745|M.smegmatis_MC2_155 VRVYLATGLTDVGRPEAEHEEADME-VARVPLKEAVARVFSGDIVNGIAV
TH_1543|M.thermoresistible__bu VRVYLATGLTEVPRPDAHHEEADLT-LRWFPLAEAVRMVLAGEIVNSIAV
MAB_2365|M.abscessus_ATCC_1997 VRVFLATGLSHPGRGESHDEEADMT-VHWVPLSEAVSMVMAGEVVNAIAV
MLBr_02698|M.leprae_Br4923 VHKTVHHYLMRFSGGELSDDDLEVTEVAWVPIRELPSRLAYADERRLAEV
*: : * : .:: :: : : : : .: . :
Mb1726|M.bovis_AF2122/97 AGVLAVHAVTTG----FAQPRPLDTEWIDRPTAFATRRAER---------
Rv1700|M.tuberculosis_H37Rv AGVLAVHAVTTG----FAQPRPLDTEWIDRPTAFAARRAER---------
MAV_3071|M.avium_104 AGILAAHAVTTG----FERPRPVDSPWQDRPTAFPARKAGR---------
MMAR_2505|M.marinum_M AGVLAAHSISTG----LAQPRPLDSPWIDRPTAFMARKNSG---------
MUL_1689|M.ulcerans_Agy99 AGVLAAHSISTG----LAQPRPLDSPWIDRPTAFMARKNSR---------
Mflv_3501|M.gilvum_PYR-GCK AGILAAHTMTG-----TDALRPVDAPWVDRPQALGRRLGHS---------
Mvan_3282|M.vanbaalenii_PYR-1 AGILAAHAMPD-----VSTLRPVDAPWVDRPTAFARRVGHP---------
MSMEG_3745|M.smegmatis_MC2_155 AGILAAHSAKD-----IESLRRVDAPWVDRPTAFWRRKGLS---------
TH_1543|M.thermoresistible__bu AGLLAAHAHAGAHPGPTAELRSVDAPWPDRPTDFAARKAHR---------
MAB_2365|M.abscessus_ATCC_1997 AGILATHIALGG-----HHTRPVDAPWPDRPTAFAERKAAT---------
MLBr_02698|M.leprae_Br4923 AYELINKLQTGG--PAALPPLPPSLPRRQSQTHFRTRHSNNSANSRKNGH
* * : . : : *
Mb1726|M.bovis_AF2122/97 ----
Rv1700|M.tuberculosis_H37Rv ----
MAV_3071|M.avium_104 ----
MMAR_2505|M.marinum_M ----
MUL_1689|M.ulcerans_Agy99 ----
Mflv_3501|M.gilvum_PYR-GCK ----
Mvan_3282|M.vanbaalenii_PYR-1 ----
MSMEG_3745|M.smegmatis_MC2_155 ----
TH_1543|M.thermoresistible__bu ----
MAB_2365|M.abscessus_ATCC_1997 ----
MLBr_02698|M.leprae_Br4923 GQGP