For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SHVQLDAIGYHLPDGRALLHDVSLRVGEGSKTALIGPNGTGKTTLLRIIAGDADPHDGAVTRGGQLGVMR QFIGSVRDDSTVRDLLLSVAPDRIRRAAEHLDRAELALMECDSEQDQIKYAQALADWADVGGYEFETQCD VHTTAALGIPYELARFRGVNTLSGGQQKRLVLESLLGGPHQVLLLDEPDNYLDVPAKRWLEERLVESPKT ILFVSHDRELINRAAGQIATLEPTRAGSTLWVHPGSFTTYHQARADRNAKLAELRRRWDEQRAALRDLVL MYRQKAAYNSDMASRLQAAETRLRRFDEAGPPEAVPLRQNVRMRLAGGRTAKRAVIADNLELTGLTRPFD TELWYGDRVAVLGGNGTGKSHFLRLLACGGTDPELDQLPVGDMIPEPVNHDGRLRLGARVRPGWFAQTHH HAGLMDRTLLDILHHGDERRAGHGREQASRILDRYGLAPSAEQTFSSLSGGQQGRFQILLLELMGSTLLL LDEPTDNLDLHSAEALEDALAAFDGTVIAVTHDRWFARTFTRFLIFGEDGKVRESVEPQWV*
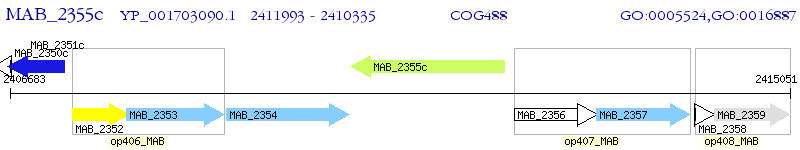
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_2355c | - | - | 100% (552) | putative ABC transporter ATP-binding protein |
| M. abscessus ATCC 19977 | MAB_2736c | - | 3e-43 | 29.50% (522) | macrolide ABC transporter ATP-binding protein |
| M. abscessus ATCC 19977 | MAB_1560 | - | 2e-40 | 27.14% (538) | putative ABC transporter ATP-binding protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1508 | - | 3e-45 | 31.13% (514) | macrolide ABC transporter ATP-binding protein |
| M. gilvum PYR-GCK | Mflv_3668 | - | 3e-44 | 30.23% (516) | ABC transporter related |
| M. tuberculosis H37Rv | Rv1473 | - | 3e-45 | 31.13% (514) | macrolide ABC transporter ATP-binding protein |
| M. leprae Br4923 | MLBr_01816 | - | 8e-44 | 30.93% (527) | ABC transporter ATP-binding protein, possibly in EF-3 subfamily |
| M. marinum M | MMAR_2279 | - | 1e-45 | 31.09% (521) | ATP-binding protein ABC transporter |
| M. avium 104 | MAV_3306 | - | 3e-46 | 30.46% (522) | ABC transporter ATP-binding protein |
| M. smegmatis MC2 155 | MSMEG_3140 | - | 1e-45 | 29.98% (517) | ABC transporter ATP-binding protein |
| M. thermoresistible (build 8) | TH_4108 | - | 8e-47 | 30.17% (527) | PUTATIVE ABC transporter related |
| M. ulcerans Agy99 | MUL_1481 | - | 6e-46 | 31.09% (521) | ATP-binding protein ABC transporter |
| M. vanbaalenii PYR-1 | Mvan_2742 | - | 4e-44 | 29.42% (520) | ABC transporter-related protein |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_2279|M.marinum_M ---------------------MITATDLEVRAG-ARILLSPDGPDLRVQP
MUL_1481|M.ulcerans_Agy99 ---------------------MITATDVEVRAG-ARILLSPDGPDLRVQP
MAV_3306|M.avium_104 ---------------------MITATDLEVRAG-ARILLSPDGPDLRVQP
Mb1508|M.bovis_AF2122/97 ---------------------MITATDLEVRAG-ARILLAPDGPDLRVQP
Rv1473|M.tuberculosis_H37Rv ---------------------MITATDLEVRAG-ARILLAPDGPDLRVQP
MLBr_01816|M.leprae_Br4923 ---------------------MITATDLEVRAG-ARILLSSDGPDLRVQP
Mflv_3668|M.gilvum_PYR-GCK ---------------------MITATDLEVRAG-ARTLLSVEGAALRVQP
Mvan_2742|M.vanbaalenii_PYR-1 MPTPAWVPVQPCHAYLTRSISVITATDLEVRAG-ARTLLSIEGSALRVQP
MSMEG_3140|M.smegmatis_MC2_155 ---------------------MITATDLEVRAG-ARTLLLIEGSALRVQP
TH_4108|M.thermoresistible__bu VIDGENKITCRTAYHRREMANLINLERVTVGYG-TRVLLHEVS--LGVAD
MAB_2355c|M.abscessus_ATCC_199 -------------------MSHVQLDAIGYHLPDGRALLHDVS--LRVGE
: : * ** . * *
MMAR_2279|M.marinum_M GDRIGLVGRNGAGKTTTLRILAGESEPYAGSVTRAG--EIGYLPQDPKEG
MUL_1481|M.ulcerans_Agy99 GDRIGLVGRNGAGKTTTLRILAGESEPYAGSVTRAG--EIGYLPQDPKEG
MAV_3306|M.avium_104 GDRIGLVGRNGAGKTTTLRILAGETEPYAGTIARSG--EIGYLPQDPKEG
Mb1508|M.bovis_AF2122/97 GDRIGLVGRNGAGKTTTLRILAGEVEPYAGSVTRAG--EIGYLPQDPKVG
Rv1473|M.tuberculosis_H37Rv GDRIGLVGRNGAGKTTTLRILAGEVEPYAGSVTRAG--EIGYLPQDPKVG
MLBr_01816|M.leprae_Br4923 GDRIGLVGRNGAGKTTTLRILAGATEPYAGSVTWTG--EIGYLPQDPKVG
Mflv_3668|M.gilvum_PYR-GCK GDRIGLVGRNGAGKTTTMRILAGEGEPYAGSVTRTG--EIGYLPQDPKEG
Mvan_2742|M.vanbaalenii_PYR-1 GDRIGLVGRNGAGKTTTMRILAGEGEPYAGSVTRTG--EIGYLPQDPKEG
MSMEG_3140|M.smegmatis_MC2_155 GDRIGLVGRNGAGKTTTMRILAGEGEPYAGSVTRSG--DIGYLPQDPKEG
TH_4108|M.thermoresistible__bu GDAIGVVGRNGDGKSTLLGVLTGEREPDSGRVTHTSGLSVGYLRQGDDFG
MAB_2355c|M.abscessus_ATCC_199 GSKTALIGPNGTGKTTLLRIIAGDADPHDGAVTRGG--QLGVMRQFIGSV
*. .::* ** **:* : :::* :* * :: . .:* : *
MMAR_2279|M.marinum_M DLEVLARDRVLSARGLDVLLTDLEKQQALMAEVADDAARDRAIRRYGQLE
MUL_1481|M.ulcerans_Agy99 DLEVLARDRVLSARGLDVLLTDLEKQQALMAEVADDAARDRAIRRYGQLE
MAV_3306|M.avium_104 DLDVLARDRVLSARGLDVLLTDLEKQQALMAEVADDDARDRAIRRYGQLE
Mb1508|M.bovis_AF2122/97 DLDVLARDRVLSARGLDVLLTDLEKQQALMAEVADEDERDRAIRRYGQLE
Rv1473|M.tuberculosis_H37Rv DLDVLARDRVLSARGLDVLLTDLEKQQALMAEVADEDERDRAIRRYGQLE
MLBr_01816|M.leprae_Br4923 NLDVLARDRVLSARGLDVLLTDLEKQQALMAEVADDDARDRAIRRYGQLE
Mflv_3668|M.gilvum_PYR-GCK DLDMLARDRVLSARGLDTLLSDLEKQQALMAEVADDAARDKAVRRYGQLE
Mvan_2742|M.vanbaalenii_PYR-1 DLDMLARDRVLSARGLDTLLSDLEKQQALMAEVADDAARDKAVRRYGQLE
MSMEG_3140|M.smegmatis_MC2_155 DLDVLARDRVLSARGLDTLLADLEKQQALMAEVADDAARDKAVRRYGQLE
TH_4108|M.thermoresistible__bu --DASIREVIVAGRPDHVWAADPTTREVVEQLLGG---------------
MAB_2355c|M.abscessus_ATCC_199 RDDSTVRDLLLSVAPDRIRRAAEHLDRAELALMECDSEQDQIKYAQALAD
: *: ::: : .. :
MMAR_2279|M.marinum_M ERFVALGGYGAESEASRICASLGLPERVLTQQLRTLSGGQRRRVELARIL
MUL_1481|M.ulcerans_Agy99 ERFVTLGGYGAESEASRICASLGLPERVLTQQLRTLSGGQRRRVELARIL
MAV_3306|M.avium_104 ERFVALGGYGAESEASRICASLGLPERVLTQQLRTLSGGQRRRVELARIL
Mb1508|M.bovis_AF2122/97 ERFVALGGYGAESEAGRICASLGLPERVLTQRLRTLSGGQRRRVELARIL
Rv1473|M.tuberculosis_H37Rv ERFVALGGYGAESEAGRICASLGLPERVLTQRLRTLSGGQRRRVELARIL
MLBr_01816|M.leprae_Br4923 ERFLALGGYSAESEAGRICASLGLPERVLVQQLCTLSGGQRRRVELARIL
Mflv_3668|M.gilvum_PYR-GCK ERFAALGGYAAESEAGRICASLGLPDRVLTQPLRTLSGGQRRRVELARIL
Mvan_2742|M.vanbaalenii_PYR-1 ERFAALGGYAAESEAGRICASLGLPDRVLTQPLRTLSGGQRRRVELARIL
MSMEG_3140|M.smegmatis_MC2_155 ERFSALGGYAAESEAGRICASLGLPERVLTQPLHTLSGGQRRRVELARIL
TH_4108|M.thermoresistible__bu --------------------------IDLDGSVDTLSGGERRRVALAAVL
MAB_2355c|M.abscessus_ATCC_199 WADVGGYEFETQCDVHTTAALGIPYELARFRGVNTLSGGQQKRLVLESLL
: *****:::*: * :*
MMAR_2279|M.marinum_M FAASD---TGAGSSTVLLLDEPTNHLDADSVGWLRDFLR----AHTGGLI
MUL_1481|M.ulcerans_Agy99 FAASD---TGAGSSTVLLLDEPTNHLDADSVGWLRDFLR----AHTGGLI
MAV_3306|M.avium_104 FAAAEGGATGSGSATTLLLDEPTNHLDADSIGWLRDFLR----AHTGGLV
Mb1508|M.bovis_AF2122/97 FAASE---SGAGNSTTLLLDEPTNHLDADSLGWLRDFLR----LHTGGLV
Rv1473|M.tuberculosis_H37Rv FAASE---SGAGNSTTLLLDEPTNHLDADSLGWLRDFLR----LHTGGLV
MLBr_01816|M.leprae_Br4923 FAASRAGTGASGSGTTLLLDEPTNHLDADSLGWLRDFLR----SHTGGLV
Mflv_3668|M.gilvum_PYR-GCK FAASE---GGGGDATTLLLDEPTNHLDADSIGWLRAFLQ----NHNGGLV
Mvan_2742|M.vanbaalenii_PYR-1 FAASE---AGSGSATTLLLDEPTNHLDADSIGWLRTFLQ----NHTGGLV
MSMEG_3140|M.smegmatis_MC2_155 FAASE---TGSGSATTLLLDEPTNHLDADSIGWLRDFLK----NHTGGLV
TH_4108|M.thermoresistible__bu IGGHD----------VLVLDEPTNHLDVEVIAWLAAHLSGRTARRTGALV
MAB_2355c|M.abscessus_ATCC_199 GGPHQ----------VLLLDEPDNYLDVPAKRWLEERLVE----SPKTIL
. .*:**** *:**. ** * ::
MMAR_2279|M.marinum_M IISHNVELLADVVNRVWFLDAVRG--EVDVYNMGWHKYLDARATDEQRRR
MUL_1481|M.ulcerans_Agy99 IISHNVELLADVVNRVWFLDAVRG--EVDVYNMGWHKYLDARATDEQRRR
MAV_3306|M.avium_104 IISHNVELLADVVNRVWFLDAVRG--EVDVYNMSWQKYLDARATDEQRRR
Mb1508|M.bovis_AF2122/97 VISHNVDLVADVVNKVWFLDAVRG--QVDVYNMGWQRYVDARATDEQRRI
Rv1473|M.tuberculosis_H37Rv VISHNVDLVADVVNKVWFLDAVRG--QVDVYNMGWQRYVDARATDEQRRI
MLBr_01816|M.leprae_Br4923 VISHNVELIAAVVNRVWFLDAVLG--KVDVYNMGWYKYLDSRATDEQRRR
Mflv_3668|M.gilvum_PYR-GCK VISHDVELLADVVNRVWFLDAVRG--EADVYNMGWQKYLDARATDEQRRR
Mvan_2742|M.vanbaalenii_PYR-1 VISHDVDLLADVVNRVWFLDAVRG--EADVYNMGWQKYLDARATDEQRRR
MSMEG_3140|M.smegmatis_MC2_155 MISHNVELLEEVVNRVWFLDAVRG--EVDVYNMGWQKYLDARATDEQRRR
TH_4108|M.thermoresistible__bu VVSHDRWFLDAVCTTTWEVHSASGKGTVDAYEGGYAAYVLARAER----M
MAB_2355c|M.abscessus_ATCC_199 FVSHDRELINRAAGQIATLEPTRAGSTLWVHPGSFTTYHQARADRNAKLA
.:**: :: . :... . .: .: * :**
MMAR_2279|M.marinum_M RERANAERKATALRTQAAKMGAKATKAVAAQNMLRRADRMMAALDEERVA
MUL_1481|M.ulcerans_Agy99 RERANAERKATALRTQAAKMGAKATKAVAAQNMLRRADRMMAALDEERVA
MAV_3306|M.avium_104 RERANAERKAAALRTQAAKLGAKATKAVAAQNMLRRADRMMAALDEERVA
Mb1508|M.bovis_AF2122/97 RERANAERKAAALRAQAAKLGAKATKAVAAQNMLRRADRMMAALDEERVA
Rv1473|M.tuberculosis_H37Rv RERANAERKAAALRAQAAKLGAKATKAVAAQNMLRRADRMMAALDEERVA
MLBr_01816|M.leprae_Br4923 RERVNAERKATALRTQAAKMGAKATKAVAAQNMLRRADRMMAALDEERVI
Mflv_3668|M.gilvum_PYR-GCK RERANAEKKAGALRAQAAKMGAKATKAVAAQNMLRRAERMMAELDAERVA
Mvan_2742|M.vanbaalenii_PYR-1 RERANAEKKASALRAQAAKMGAKATKAVAAQNMLKRAERMMAALDDERVA
MSMEG_3140|M.smegmatis_MC2_155 RERANAEKKAGALRAQAAKMGAKATKAVAAQNMLRRAERMMAALDEERVA
TH_4108|M.thermoresistible__bu RLAATAETRRRNLLRKELAWLRRGPPARTSKPKFRIQAANELIATEPPPR
MAB_2355c|M.abscessus_ATCC_199 ELRRRWDEQRAALRDLVLMYRQKAAYNSDMASRLQAAETRLRRFDEAGPP
. : : * :.. ::
MMAR_2279|M.marinum_M DKVARIKFPTPAACGRVPLVVKGLSKTYG--SLEVFTGVDLAIDRGSRVV
MUL_1481|M.ulcerans_Agy99 DKVARIKFPTPAACGRVPLVVKGLSKTYG--SLEVFTGVDLAIDRGSRVV
MAV_3306|M.avium_104 DKVARIKFPTPAACGRTPLTATGLSKSYG--SLEVFSGVDLAIDRGSRVV
Mb1508|M.bovis_AF2122/97 DKVARIKFPTPAACGRTPLVANGLGKTYG--SLEVFTGVDLAIDRGSRVV
Rv1473|M.tuberculosis_H37Rv DKVARIKFPTPAACGRTPLVANGLGKTYG--SLEVFTGVDLAIDRGSRVV
MLBr_01816|M.leprae_Br4923 EKVAQIKFPTPAACGRTPLVAKGLSKTYG--SLEVFTGVNLAIDRGSRVV
Mflv_3668|M.gilvum_PYR-GCK DKVARIKFPTPAPCGKTPLIAKGLTKTYG--SLEIFTGLDLAIDRGSRVV
Mvan_2742|M.vanbaalenii_PYR-1 DKVARIKFPTPAPCGKTPLTAKGLTKTYG--SLEIFSGLDLAIDRGSRVV
MSMEG_3140|M.smegmatis_MC2_155 DKVARIKFPTPAPCGRTPLVAKGLTKNYG--SLEVFTGVDLAIDRGSRVV
TH_4108|M.thermoresistible__bu DSVVLQRFAT-SRLGKDVFDLHGVRLEVGEPPRVVLDNIDWSIGPGDRIG
MAB_2355c|M.abscessus_ATCC_199 EAVPLRQNVRMRLAGGRTAKRAVIADNLE--LTGLTRPFDTELWYGDRVA
: * : * : : .: : *.*:
MMAR_2279|M.marinum_M VLGLNGAGKTTLLRLLAG-----------------VEHPDTGGLEPGHGL
MUL_1481|M.ulcerans_Agy99 VLGLNGAGKTTLLRLLAG-----------------VEHPDTGGLEPGHGL
MAV_3306|M.avium_104 VLGLNGAGKTTLLRLLAG-----------------AETPDTGRIEPGHGL
Mb1508|M.bovis_AF2122/97 ILGLNGAGKTTLLRLLAG-----------------VEQPDTGVLEPGYGL
Rv1473|M.tuberculosis_H37Rv ILGLNGAGKTTLLRLLAG-----------------VEQPDTGVLEPGYGL
MLBr_01816|M.leprae_Br4923 VLGLNGAGKTTLLRVLAG-----------------AEKPDVGRLEPGHGL
Mflv_3668|M.gilvum_PYR-GCK VLGLNGAGKTTLLRLLAG-----------------VEIADAGGLEPGHGL
Mvan_2742|M.vanbaalenii_PYR-1 VLGLNGAGKTTLLRLLAG-----------------VEKADAGEIEPGHGL
MSMEG_3140|M.smegmatis_MC2_155 VLGLNGAGKTTLLRLIAG-----------------TEKPDAGHLEPGHGL
TH_4108|M.thermoresistible__bu LVGVNGTGKTSVLRMLAG-----------------QLQPTAGTVKRGATL
MAB_2355c|M.abscessus_ATCC_199 VLGGNGTGKSHFLRLLACGGTDPELDQLPVGDMIPEPVNHDGRLRLGARV
::* **:**: .**::* * :. * :
MMAR_2279|M.marinum_M RIGYFAQEHDTLDNDATVWENIR-----HAAPESGEQELRGLLGAFMFTG
MUL_1481|M.ulcerans_Agy99 RIGYFAQEHDTLDNDATVWENIR-----HAAPESGEQELRGLLGAFMFTG
MAV_3306|M.avium_104 RIGYFAQEHDTLDNDATVWQNIR-----HAAPDSGEQDLRGLLGAFMFSG
Mb1508|M.bovis_AF2122/97 RIGYFAQEHDTLDNDATVWENVR-----HAAPDAGEQDLRGLLGAFMFTG
Rv1473|M.tuberculosis_H37Rv RIGYFAQEHDTLDNDATVWENVR-----HAAPDAGEQDLRGLLGAFMFTG
MLBr_01816|M.leprae_Br4923 RIGYFAQEHDTLDNEDTVWQNIR-----HTAPEFGEQDLRGLLGAFMFSG
Mflv_3668|M.gilvum_PYR-GCK KIGYFAQEHDTLDTMRTVWENIR-----SAAPDTGEQDLRGLLGAFMFTG
Mvan_2742|M.vanbaalenii_PYR-1 KIGYFAQEHDTLDNMASVWENIR-----HAAPDTGEQDLRGLLGAFMFTG
MSMEG_3140|M.smegmatis_MC2_155 KIGYFAQEHDTLDNQATVWENIR-----HAAPDTGEQDLRGLLGAFMFTG
TH_4108|M.thermoresistible__bu KIGYLSQALDELDGSDRVLDAVENRRRITELAGGREISAATLLEDFGFTG
MAB_2355c|M.abscessus_ATCC_199 RPGWFAQTHHHAGLMDRTLLDILHHGD-ERRAGHGREQASRILDRYGLA-
: *:::* . . . : . . . :* : ::
MMAR_2279|M.marinum_M PQLDQPAGTLSGGEKTRLALAGLVASTANVLLLDEPTNNLDPASREQVLD
MUL_1481|M.ulcerans_Agy99 PQLDQPAGTLSGGEKTRLALAGLVASTANVLLLDEPTNNLDPASREQVLD
MAV_3306|M.avium_104 PQLDQPAGTLSGGEKTRLALAGLVASTANVLLLDEPTNNLDPASREQVLD
Mb1508|M.bovis_AF2122/97 PQLEQPAGTLSGGEKTRLALAGLVASTANVLLLDEPTNNLDPASREQVLD
Rv1473|M.tuberculosis_H37Rv PQLEQPAGTLSGGEKTRLALAGLVASTANVLLLDEPTNNLDPASREQVLD
MLBr_01816|M.leprae_Br4923 PQLDQLAGTLSGGEKTRLALAGLVASTANVLLLDEPTNNLDPASRKQVLD
Mflv_3668|M.gilvum_PYR-GCK PQLEQPAGTLSGGEKTRLALAGLVASTANVLLLDEPTNNLDPASREQVLD
Mvan_2742|M.vanbaalenii_PYR-1 PQLDQPAGTLSGGEKTRLALAGLVASTANVLLLDEPTNNLDPASREQVLD
MSMEG_3140|M.smegmatis_MC2_155 PQLDQPAGTLSGGEKTRLALAGLVASTANVLLLDEPTNNLDPASREQVLD
TH_4108|M.thermoresistible__bu DKLTTRIADLSGGERRRLQFLRLLLDEPNVLLLDEPTNDLDIDTLTVIED
MAB_2355c|M.abscessus_ATCC_199 PSAEQTFSSLSGGQQGRFQILLLELMGSTLLLLDEPTDNLDLHSAEALED
. . ****:: *: : * ..:*******::** : : *
MMAR_2279|M.marinum_M ALRSYQGAVVLVTHDPGAAEALDPQ--------RVVLLPDATEDYWSDEY
MUL_1481|M.ulcerans_Agy99 ALRSYQGAVVLVTHDPGAAEALDPQ--------RVVLLPDATEDYWSDEY
MAV_3306|M.avium_104 ALRSYTGAVVLVTHDPGAAEALDPQ--------RVVLLPDGTEDYWSDEY
Mb1508|M.bovis_AF2122/97 ALRSYRGAVVLVTHDPGAAAALGPQ--------RVVLLPDGTEDYWSDEY
Rv1473|M.tuberculosis_H37Rv ALRSYRGAVVLVTHDPGAAAALGPQ--------RVVLLPDGTEDYWSDEY
MLBr_01816|M.leprae_Br4923 ALRNYQGAVVLVTHDPGAAKALDPQ--------RVVLLPDGTEDYWSDEY
Mflv_3668|M.gilvum_PYR-GCK ALRSYAGAVVLVTHDPGAAEALDPQ--------RVLLLPDGTEDFWSDDY
Mvan_2742|M.vanbaalenii_PYR-1 ALRSYVGSVVLVTHDPGAAEALDPQ--------RVLLLPDGTEDFWSDEY
MSMEG_3140|M.smegmatis_MC2_155 ALRSYEGAVVLVTHDPGAAEALDPQ--------RVVLLPDGTEDFWSDEY
TH_4108|M.thermoresistible__bu YLDGWPGTVIVVTHDRYFLERVSDVTYALTGGGRCDLLPGGVDQYLSDRA
MAB_2355c|M.abscessus_ATCC_199 ALAAFDGTVIAVTHDRWFARTFTRF------------LIFGEDGKVRESV
* : *:*: **** . * . : :
MMAR_2279|M.marinum_M RDLIELA-------------------------------------------
MUL_1481|M.ulcerans_Agy99 RDLIELA-------------------------------------------
MAV_3306|M.avium_104 RDLIELA-------------------------------------------
Mb1508|M.bovis_AF2122/97 RDLIELA-------------------------------------------
Rv1473|M.tuberculosis_H37Rv RDLIELA-------------------------------------------
MLBr_01816|M.leprae_Br4923 RDLIELA-------------------------------------------
Mflv_3668|M.gilvum_PYR-GCK RDLIELA-------------------------------------------
Mvan_2742|M.vanbaalenii_PYR-1 RDLIELA-------------------------------------------
MSMEG_3140|M.smegmatis_MC2_155 RDLIELA-------------------------------------------
TH_4108|M.thermoresistible__bu GDRAARKAPTAKSPAATGAGAGKGESPAARQRRIAKELTRIEGQLSKIDG
MAB_2355c|M.abscessus_ATCC_199 EPQWV---------------------------------------------
MMAR_2279|M.marinum_M ----------------------------------------------
MUL_1481|M.ulcerans_Agy99 ----------------------------------------------
MAV_3306|M.avium_104 ----------------------------------------------
Mb1508|M.bovis_AF2122/97 ----------------------------------------------
Rv1473|M.tuberculosis_H37Rv ----------------------------------------------
MLBr_01816|M.leprae_Br4923 ----------------------------------------------
Mflv_3668|M.gilvum_PYR-GCK ----------------------------------------------
Mvan_2742|M.vanbaalenii_PYR-1 ----------------------------------------------
MSMEG_3140|M.smegmatis_MC2_155 ----------------------------------------------
TH_4108|M.thermoresistible__bu QIAALHEAMAAAASDHVRLAELNTELGELQRRKTRLEEDWLITADG
MAB_2355c|M.abscessus_ATCC_199 ----------------------------------------------